Umweltmikrobiologie und Biotechnologie (UMB)
Herzlich willkommen
auf der Internetpräsenz des Umweltmikrobiologie und Biotechnologie (UMB).
Wir möchten Sie hier über die Forschung in den Arbeitsgruppen Aquatische Mikrobiologie und Molekulare Enzymtechnologie und Biochemie informieren.
Sie erhalten außerdem Informationen über aktuelle Projekte und Publikationen sowie die Mitarbeiter der einzelnen Arbeitsgruppen.
Studierende finden hier mögliche Themen für Bachelor- und Masterarbeiten.
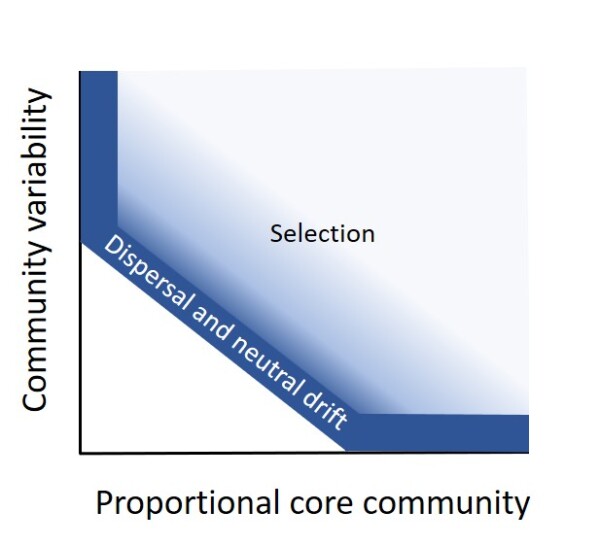
09.04.2024 New Paper: Imprints of ecological processes in the taxonomic core community: an analysis of naturally replicated microbial communities enclosed in oil
It is widely assumed that a taxonomic core community emerges among microbial communities from similar habitats because similar environments select for the same taxa bearing the same traits. Yet, a core community itself is no indicator of selection because it may also arise from dispersal and neutral drift, i.e. by chance. Here, we hypothesize that a core community produced by either selection or chance processes should be distinguishable. While dispersal and drift should produce core communities with similar relative taxon abundances, especially when the proportional core community, i.e. the sum of the relative abundances of the core taxa, is large, selection may produce variable relative abundances. We analyzed the core community of 16S rRNA gene sequences of 193 microbial communities occurring in tiny water droplets enclosed in heavy oil from the Pitch Lake, Trinidad and Tobago. These communities revealed highly variable relative abundances along with a large proportional core community (68.0 ± 19.9 %). A dispersal-drift null model predicted a negative relationship of proportional core community and compositional variability along a range of dispersal probabilities and was largely inconsistent with the observed data, suggesting a major role of selection for shaping the water droplet communities in the Pitch Lake.
01.03.2024 Neue Doktorandinnen in der aquatischen Mikrobiologie
Zwei neue Doktorandinnen in der aquatischen Mikrobiologie: Frau Janina Oldemeyer untersucht neue Schlüsselenzyme im anaeroben Abbau von Naphthalin und Frau Isabell Erdmann entwickelt einen mikrobiellen Index um den Ökosystemstatus von Grundwasserkörpern bestimmen zu können.

December 2023 New Funding by Fonds der Chemischen Industrie (FCI)
The junior research group on Microbial Ecotones lead by Lisa Voskuhl is pleased to announce that the Fonds der Chemischen Industrie has awarded us a grant for material costs for the project "Extraction and analysis of alkanes and (poly)aromatic hydrocarbon compounds from algae".

01.02.2023 New Junior Research Group on Microbiology of Ecotones
Lisa Voskuhl established a new junior research group at the Depatment of Envrionmenal Microbiology and Biotechnology (EMB)
The group is interested in studying microbial processes in environmentally relevant ecotones, particularly oil/water and algae phycosphere/water.
Jäger der verborgenen Schätze
Zwischen Bären und heißen Quellen: Im Rahmen des deutsch-russischen DFG-RSF Kooperationsprojektes ExploCarb (Life Science Programm) hat ein fünfköpfiges Team aus Mikrobiologen zwei Wochen in der Uson-Caldera in Kamschatka verbracht – unter ihnen ein Nachwuchsforscher der UDE. In dieser Zeit suchten sie mithilfe sowohl bewährter, als auch neuer an der UDE etablierter Methoden nach bisher unentdeckten Mikroorganismen und Enzymen.
Mit einer Vielzahl an heißen Quellen, Fumerolen und Geysiren bietet der Krater des ehemaligen Vulkans Uson ideale Bedingungen für Mikroorganismen, die sich auf extreme Bedingungen spezialisiert haben. Auffallend häufig anzutreffen sind in solchen Habitaten Vertreter aus der Domäne der Archaea. Diese einzelligen Lebewesen weisen einzigartige stoffwechselphysiologischen Eigenschaften auf, weshalb sie für die anwendungsorientierte als auch für die Grundlagenforschung höchst interessant sind.
Ziel des Forschungsprojektes ExploCarb, an dem sich neben UDE-Forschern aus den Gruppen von Bettina Siebers (Molekulare Enzymtechnologie und Biochemie) und Markus Kaiser (Chemische Biologie) auch Wissenschaftler der Russischen Akademie der Wissenschaften  (Ilya Kublanov, Extremophiles Metabolism Laboratory) beteiligen, ist es, den Kohlenstoffwechsel in besonders hitzeliebenden („hyperthermophilen“) Archaeen zu erforschen, die bei Temperaturen über 80°C wachsen, und neue interessante Hydrolasen für die Biotechnologie zu gewinnen.
(Ilya Kublanov, Extremophiles Metabolism Laboratory) beteiligen, ist es, den Kohlenstoffwechsel in besonders hitzeliebenden („hyperthermophilen“) Archaeen zu erforschen, die bei Temperaturen über 80°C wachsen, und neue interessante Hydrolasen für die Biotechnologie zu gewinnen.
von links nach rechts: Kseniya Zayulina, Alexander Elcheninov, Thomas Klaus, Dr Tatjana Kochetkova, Dr Alexandra Popova
Hierbei soll auch erstmals die von den Kollegen der UDE etablierte Methode des aktivitäts-basierten Protein-Profiling (ABPP, doi: 10.1038/ncomms15352) zur Identifizierung neuer Enzyme-eingesetzt werden. Darüber hinaus soll das Projekt auch dazu beitragen den interkulturellen und wissenschaftlichen Austausch zwischen beiden Ländern zu fördern – darüber, dass dies insbesondere durch die gemeinsame Expedition sowie Projekttreffen bereits sehr erfolgreich gelungen ist, sind sich alle Beteiligten einig.
Activity-based protein profiling as a robust method for enzyme identification and screening in extremophilic Archaea. Zweerink S, Kallnik V, Ninck S, Nickel S, Verheyen J, Blum M, Wagner A, Feldmann I, Sickmann A, Albers SV, Bräsen C, Kaschani F, Siebers B, Kaiser M. Nat Commun. 2017 May 8;8:15352. doi: 10.1038/ncomms15352. PMID: 28480883

Laura Kuschmierz gewinnt Poster Preis der GRC
Laura Kuschmierz hat auf der Gordon Research Conference on Archaea 2019 in Les Diablerets (Schweiz) einen von zwei Posterpreisen gewonnen. Das Poster trug den Titel : "Archaeal Biofilms: Composition of extracellular polymeric substances, exopolysaccharide synthesis and secretion in Sulfolobus acidocaldarius."

Ankündigung: Hans-Curt Flemming Lecture
Zu Ehren des ehemaligen und emeritierten Lehrstuhlinhabers, Prof. Dr. Hans-Curt Flemming, hat das Biofilm Center absofort die Hans-Curt Flemming Lecture ins Leben gerufen. In der Vortragsreihe werden unter anderem nationale und internationale Spitzenwissenschaftler Vorträge halten. Das Vortragsreihe ersetzt das bisherige Biofilm Center Seminar und findet momentan Montags 14 Uhr in T03 R03 D89 statt.
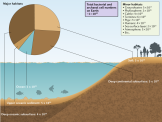
Forscher des Biofilm-Centers klären auf: Wie viele Mikroben gibt es auf unserem Planeten und was ist ihre bevorzugte Lebensweise?
Biofilme stellen eine Form kollektiven Lebens mit Eigenschaften dar, die viele Vorteile für ihre Bewohner bietet und eine deutlich höhere Ebene der Organisation darstellt als das Leben von Einzelzellen. Bis heute weiß man aber nicht, wie groß der Anteil an Biofilmen, global gesehen, tatsächlich ist. Hans-Curt Flemming und Stefan Wuertz haben die neuesten Zahlen zusammengetragen (wohl eingedenk der Unsicherheiten). Die meisten Bakterien und Archäen auf der Erde (1,2 x 1030) existieren in den “Big Five Habitats”: tiefe (4 x 1029) und obere (5 x 1028) Meeressedimente, kontinentaler Tiefenbereich (3 x 1029), Böden (3 x 1029) sowie Ozeane (1 x 1029) Zellen. Die übrigen Habitate wie Grundwasser, Schwimmschicht der Ozeane, Menschen, Tiere, Atmosphäre und Phyllosphäre enthalten jeweils um mehrere Größenordnungen weniger Zellen. Biofilme dominieren in allen Habitaten auf der Erdoberfläche, außer den Ozeanen, und umfassen hier 80 % der prokaryotischen Zellen. Im tiefen Untergrund lassen sich jedoch einzelne sessile Zellen und Biofilme schlecht unterscheiden; die Autoren schätzen, das hier 20-80 % in Biofilmen leben, diese Biofilme jedoch die globalen biogeochemischen Prozesse betreiben. Also leben insgesamt zwischen 40 und 80 % aller Prokaryoten in Biofilmen und bilden die Hauptform aktiven mikrobiellen Lebens auf der Erde.
Originalartikel: Flemming, H.-C., Wuertz, S. (2019): Bacteria and Archaea on Earth and their abundance in biofilms. Nat. Rev. Microbiol. 17, DOI: 10.1038/s41579-019-0158-9 LESEN.

REGROUND project is featured in media
REGROUND project is featured in different platforms. A documentray film produced by the consortium lead by the members of AG Prof. R. Meckenstock was presented in Campus Essen at 29.01.2019. The film showed how the heavy metals are distributed in the environment, how REGROUND project offers a novel innovation (ColFerroX technology) to immobilize these toxic elements. The team were then interviewed by Deutschlandfunk radio. An article and a podcast can be found via this link.

Biolfim Centre participates in Wissensnacht Ruhr 2018
“WissensNachtRuhr” (Science Night Ruhr) is one of the largest Science Festivals in Germany, which takes place every two years. During this evening, scientists from institutions of the Ruhr area offer diverse activities directed to families, students and everybody interested in research and science. This year, “WissesnachtRuhr” took place on the 28th of September and the Aquatic Microbiology department was present.
Our very own team of science communicators, Ivana Kraiselburd, Meike Arnold, Ianina Kaplieva-Dudeck and Lisa Kroll hosted an interactive stand at “Haus der Technik” in Essen, where visitors were able to extract DNA, observe microorganism under the microscope and discover which microbes are growing in their skin and in every-day objects like phones.

Larissa Schocke gewinnt Posterpreis der ISE
Larissa Schocke hat auf der Extremophiles Conference 2018 in Ischia, Italien, den Posterpreis der International Society for Extremophiles gewonnen. Das Poster trug den Titel "Impact of protein phosphorylation in the thermoacidophilic crenarchaeota Sulfolobus acidocaldarius DSM 639".

14.08.2018 Two prizes for nanoremediation laboratory
The nanoremediation laboratory of Prof. Dr. R. Meckenstock recently has won two prestigious prizes. On July 13th, at the graduation ceremony of the ninth round of the founding competition of GRIID (GründungsInitiative Innovation Duisburg), Dr. Beate Krok, together with Dr. Sadjad Mohammadian and Prof. Dr. Rainer Meckenstock, won the first prize for the best business plan. The business plan was developed during the Small Businiess Management (SBM) course.
In addition, REGROUND project has been awarded to attend first phase of the EIT Raw Materials' accelerator program. The team will receive €15K as a funding to cover their costs for participating in this Accelerator Program from September to December 2018.
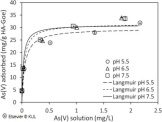
15.07.2018 New paper on Arsenic removal from Groundwater
In the framework of REGROUND project and in collaboration with the Katholieke Universiteit Leuven (KUL) and Friedrich-Schiller-Universität Jena (FSU-Jena) a new paper is published. The paper present a quantitiave analysis on the effectiveness of Humic-acid coated goethite nanoparticle in removal of Arsenic from contaminated groundwater. The paper can be accessed from this link.

02.02.2018 Hundreds of novel organisms from the deep subsurface
Several hundred novel microorganisms were discovered in groundwater fluids sourced from the deep terrestrial subsurface of the Colorado Plateau. Lead by Alexander Probst (now the head of Group of Aquatic Microbial Ecology), scientists of the University of California, Berkeley, collaborated with researchers from the University of Calgary and the Joint Genome Institute to apply microbial source tracking based on hydrogeological measurements. They discovered a stratification of microbial metabolism and diversity throughout the subsurface and postulated a symbiotic interaction between two uncultivated archaea that comprise most of the biomass in this subsurface system.
Read more here.

15.09.2017 Alexander Probst kommt ans Biofilm Centre
Der Biologe Dr. Alexander Probst wechselte von der University of California, Berkely, ans Biofilm Centre, wo er nun die die Professur für „Aquatische Mikrobielle Ökologie“ vertritt. Die Pressestelle der Universität Duisburg-Essen berichtet.

06.04.2017 Philip Weyrauch gewinnt Posterpreis
Philip Weyrauch hat auf der Microbiology Society Annual Conference 2017 in Edinburgh, GB, den Microbiology Posterpreis für das beste wissenschaftliche Poster in der Kategorie "Environmental and Applied Microbiology Forum" gewonnen. Das Poster trug den Titel "Elucidation of a dearomatising reductase reaction involved in anaerobic degradation of naphthalene".
"In the presented work we elucidated the 5,6,7,8-tetrahydro-2-naphthoyl-CoA reductase reaction in cell free extracts of sulphate-reducing naphthalene degraders by testing different electron donors. Our data indicate that the reduced ferredoxins required for this reaction are delivered via a 2-oxoglutarate:ferredoxin oxidoreductase and that the downstream pathway proceeds via β-oxidation-like reactions with water addition to hexahydro-2-naphthoyl-CoA."
23.03.2017 Doktorarbeit erfolgreich verteidigt
Am 23.03.2017 hat Hubert Müller erfolgreich seine Doktorarbeit mit dem Titel "Long-distance electron transfer by cable bacteria in aquifer sediments" verteidigt.
01.12.2016 Doktorarbeit erfolgreich verteidigt
Am 01.12.2016 hat Philip Weyrauch erfolgreich seine Doktorarbeit mit dem Titel "Mikrobieller Abbau von Naphthalin unter anaeroben Bedingungen" verteidigt.


