CRC 1093 - Project Area A
Project Areas
Area A: Chemical Ligands and Methods
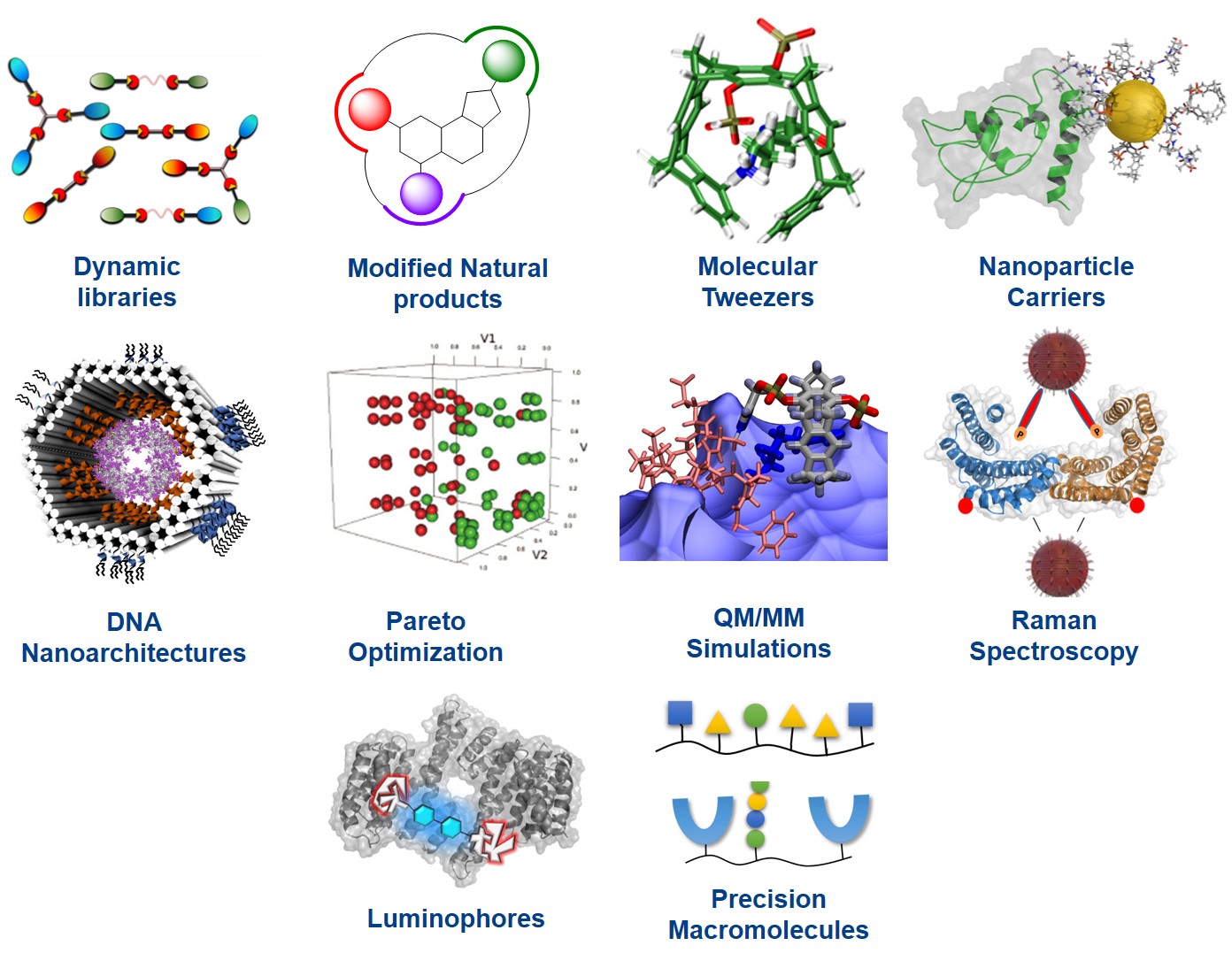
Project Area A contains all those groups involved in chemical synthesis, theoretical treatment and analytical detection of supramolecular protein ligands and their complexes. Combinatorial approaches are mainly found in projects A1-A2: A1 (Schmuck) produces focused libraries of linear and multi-armed ligands, both on the solid phase as well as with dynamic combinatorial chemistry. These also contain tailored binding sites and are screened for maximum interaction with the desired protein target. A2 (Kaiser) uses molecular docking to render unspecific naturally occurring protein ligands specific for a given protein, by filling its specificity pockets with additional synthetic moieties; it also develop self-assembled ditopic protein ligands from dynamic combinatorial libraries.
The successive projects A3, A5 and A6 describe approaches with a high degree of rational design: A3 (Schrader) provides supramolecular copolymers built from amino acid-selective monomers. For large protein assemblies imprinted nanogel materials will be constructed, which may cover the surface of the whole assembly. In addition, a family of molecular lysine and arginine tweezers is equipped with additional recognition units for specific protein surface recognition. Various tweezers will also be fused on preformed skeletons such as precision oligomers and ultrasmall nanoparticles, equipped with novel fluorophors. Group A5 (Epple) focuses on the synthesis and surface functionalization of ultrasmall gold nanoparticles with a diameter of 1-2 nm. These will be covalently functionalized by orthogonal chemical coupling (click reactions, peptide binding via cysteine) with synthetic receptor molecules and protein-specific peptides. The expertise from A6 (Saccà) aims at developing a general host-guest complexation strategy for the manipulation of the stability and activity of desired proteins specifically encapsulated into customized DNA cages. Spatial control of multivalent ligand-protein interactions will be also employed to mimic the architecture of fundamental components of the kinetochore-microtubule interface.
All supramolecular ligands are calculated and optimized for most efficient and specific protein recognition by A7 (Hoffmann) and A8 (Sanchez Garcia), which carry out Pareto optimization of their heteroavidity, with a special emphasis on solvent and entropy effects, and combine MD simulations of the whole protein/ligand complex with QM/MM calculations of the critical interface. A9 (Schlücker) pioneers the development of analytical techniques based on Raman spectroscopy which are introduced to characterize protein recognition events. Project A10 (Voskuhl) deals with the synthesis, characterisation and application of novel luminophores with aggregation-induced emission for the recognition of proteins. Finally sequence-controlled oligo(amidoamines) are applied in A11 (Hartmann) as a new platform for the multivalent presentation of non-natural supramolecular binding motifs in combination with biological ligands and sensor molecules.