Research
Central aim of our research is to contribute to a better understanding of lipids and metabolites in biological, physiological and pathophysiological processes to help in the development of a more personalized medicine as well as for a better understanding of biological processes. Therefore, we develop and apply novel and innovative workflows to identify and quantify lipids and metabolites in different samples such as plasma, serum or cells. In addition, we are apply these strategies to complex food samples for insights into the composition.
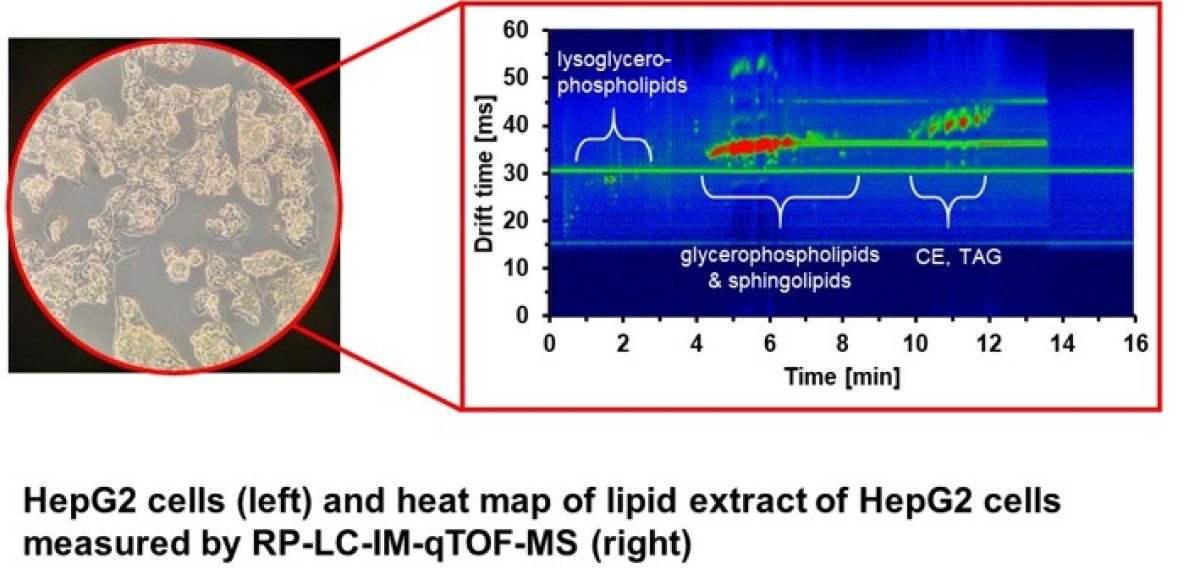
Quantitative characterization of the lipid and metabolite content of HepG2 cells
Biological samples such as plasma, serum or cells can contain hundreds or even thousands of different lipids and metabolites in various concentration, which is leading to a series of problems such as ion suppression or difficulties in the separate detection of isobaric molecules. One approach that we are following to solve these problems is to increase the separation power of the analytical platform prior to high-resolution mass spectrometric detection by using ion mobility. Liquid chromatography coupled with ion mobility quadrupole Time-of-Flight mass spectrometry (LC-IM-qTOF-MS) is especially promising to maximize the analytical coverage by separating coeluting isobaric lipid species. Besides, the mobility is characteristic for a particular ion and can be used as an additional parameter for the identification. In combination with a complete bioinformatics data analysis, this workflow enables us to analyse and identify several hundreds of lipids in different biological samples such as HepG2 cells.
(Responsible PhD.-student Kristina Rentmeister; Related funding: Metabolomics und Lipidomics 4.0: Entwicklung multidimensionaler Trenn- und Detektionstechniken zur detaillierten Charakterisierung des Metaboloms und Lipidoms biologischer Proben from the Science Support Centre of the University of Duisburg-Essen; 2018-2019)
Analysis of lipid metabolic pathways in Archaea
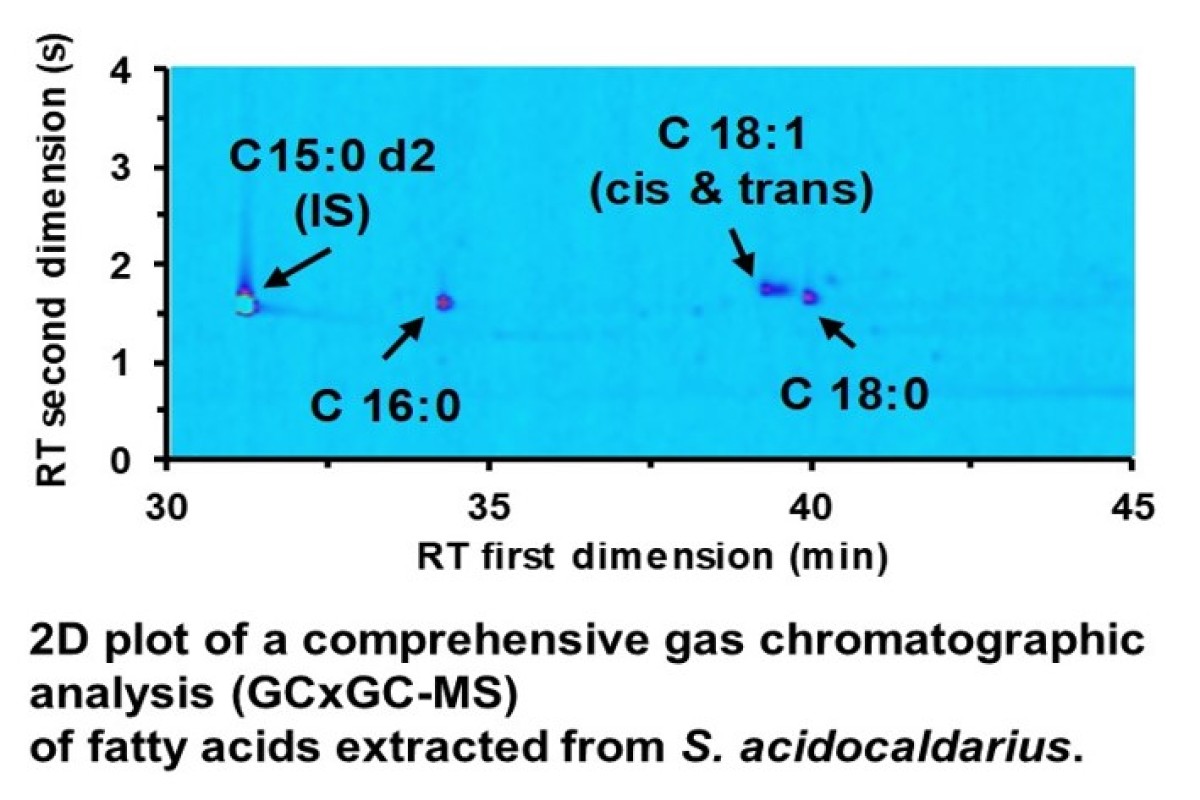
The evolution of early life into three different domains knowns as Bacteria, Archaea and Eukaryotes is yet not well understood. Recent findings showed that Eukaryotes such as humans, however, evolved from within the Archaea leading to a revised “Two domain tree of Life” model. Considering that cell membranes of Archaea are build up from isoprenoid chains ether-linked glycerol-1-phosphate lipids and comparing this to Bacteria and Eukaryotes that have membranes build up from fatty acids ester-linked glycerol-3-phosphate, a fundamental change in the membrane composition during the evolution of Archaea to Eukaryotes must have happened. How and why this “lipid divide” occurred is subject to a joined research project. For the analytical characterization of the lipid metabolic pathway -especially fatty acids and corresponding CoA-ester- we are developing and using targeted and non-targeted omics workflows.
(Responsible PhD.-student Paul Görs; Related funding: Resolving the ´lipid divide´ by unravelling the evolution and role of fatty acid metabolic pathways in Archaea – Lipid Divide from the VolkswagenStiftung; Project partners: Prof. Dr. B. Siebers, Dr. C. Bräsens, Prof. Dr. M. Kaiser University of Duisburg-Essen and Prof. Dr. T. Ettema Wageningen University NLD; 2019-2024)
Food Metabolomics: qualitative and quantitative analysis of bioactive compounds
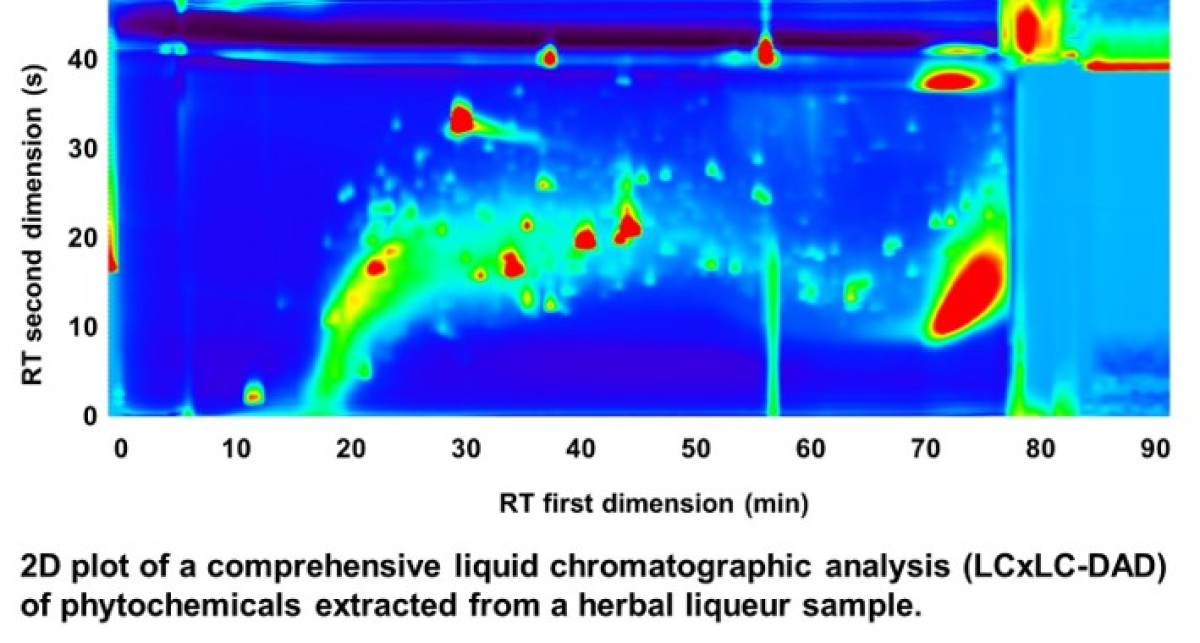
The emerging field of food metabolomics aims at the comprehensive characterization of all compounds in a food sample – similar to other omics disciplines – for insights into the composition, to understand underlying processes such as fermentation, or to confirm the authenticity. Considering that food is usually a processed mixture of plant or animal products the resulting metabolome is more complex when compared to individual ingredients. Accordingly, for a compositional characterization sophisticated analytical methods are needed. In our research, we are focussing on plant-derived food, which is known to contain biological active phytochemicals like flavonoids, stilbenoids or glucosinolate that can interact with the human metabolism. Using hyphenated techniques such as comprehensive LCxLC-DAD-MS and LC‑IM‑qTOF‑MS we are aiming to characterize complex food sample such as herbal liqueurs. These samples can be extremely complex as they are typically made by maceration of dozens of different herbs and spices. Currently we are developing workflows that are able to identify and quantify compounds from complex mixtures to allow a compositional characterization.