Complex Samples
Species- and Product Analysis
Complex samples in the field of metabolomics, lipidomics, environmental and food analysis are often analyzed by liquid chromatography coupled to a high-resolution mass spectrometer (HRMS) or by gas chromatography coupled with quadrupole-mass spectrometer (GC-MS) and the help of EI-data bases. But there is one major problem in the analysis of complex samples with LC-MS or GC-MS, which makes an analysis very difficult. With LC-MS and a high-resolution mass spectrometer (HRMS) a fantastic mass resolution with high accurate mass is possible, which allow us to calculate a sum formula. Unfortunately, it is not possible to construct the structure only with the sum formula and usually more as one, often tens or sometimes hundreds of compounds show the same sum formula, which do not allow an identification of the analytes in the sample. This is demonstrated in the figure with an ultra-high resolution FT-ICR-MS system. The m/z value of 198.18492 (figure, left top) leads to the sum formula C12H23NO (< 1.5 ppm) but cannot help us in the identification of the structure due to so many isobaric compounds (figure, right top). Also the combination with a separation method such as comprehensive two-dimensional liquid chromatography (LCxLC) in combination with a qTOF-MS (figure, below) cannot solve the problem because now, several sum formula (due to the lower mass resolution of a TOF-MS) for each spot (or peak) prevents an identification. The coupling of LC with FT-ICR-MS is not only very expensive but also not possible due to the long transient time of the FT-ICR-MS to realize a ultra-high mass resolution.
With GC-MS and the EI-data bases an identification of compounds listed in the data base is often possible, but unfortunately a lot of e.g. human or plant metabolites or environmental pollutants are not listed in these data bases. In 2015 da Silvia et al. published in PNAS that only 1.8% of spectra in an untargeted metabolomics experiment can be annotated.
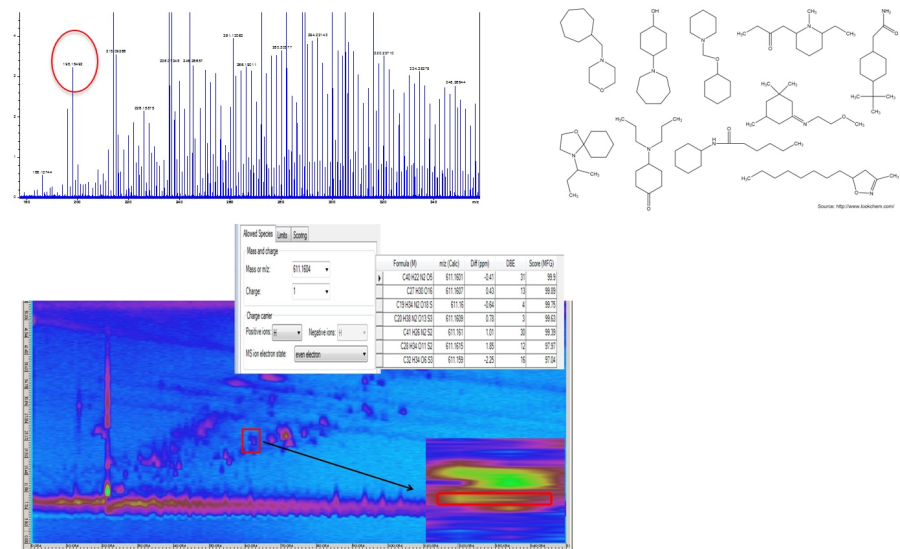
Figure1: Problems with the identification of analytes with FT-ICR-MS (top) and LCxLC-qTOF-MS (below)
This means that the vast majority of information collected by metabolomics is "dark matter", chemical signatures that remain uncharacterized. There are more than 60 million molecules in Pub Chem, yet only 220,000 MS/MS spectra representing about 20,000 molecules that are accessible for untargeted metabolomics experiments. That means, there is a serious problem in non-target analysis and we have to solve this problem due to the coming challenges in personalized medicine and environmental screening. We are working in this field to find a solution for the non-targed analysis of complex samples.

