Projects

CRC RESIST - SFB 1439
Subprojects in RESIST – a Collaborative Research Center on multiple stressors affecting streams and rivers

RESIST – a Collaborative Research Centre on multiple stressors affecting streams and rivers
Stream and rivers are degraded by a multitude of stressors – various types of pollution, degradation of hydrology and morphology, invasive species ... just to mention a few.
Stressors affect all organism groups and the functions and services they provide.
Often, stressors interact: Sometimes they strengthen each other, sometimes they weaken each other. Why and how is not fully understood. But stressor interactions greatly affects ecosystem management.
Sometimes, if a river is restored, the biota deteriorate rather than improve.
RESIST aims to unravel the interactions of multiple stressors in rivers. With manipulative lab and field experiments, field studies and models we will test why, when and how stressors interact – and develop options for mitigation and restoration.
Subproject A04: “The roles of bacteria and fungi for CPOM degradation during stressor increase and release: A metatranscriptomic approach”
The subproject A04 focuses on the role of different fungal and bacterial groups in enzymatic decomposition of coarse particulate organic matter (CPOM) - leaf decompositions - in the presence and absence of stressors. To determine the stressor effect on molecular level we will use metatranscriptome sequencing. In two experimental systems experiments will be run in collaboration with all CRC members. Here we will evaluate the effect of stressor increase and release for both fungi and bacteria. In particular, we will test whether functions recover faster than community composition to a pre‐degradation stage, due to partial functional redundancy among taxa. We expect that functions are shared between bacteria and fungi, and among different taxa within these microbial groups. Thus, if species or other taxonomic groups disappear following stressor exposure, their metabolic activity will be compensated for by other taxa. However, such compensatory mechanisms might be limited if a succession of taxa is necessary for the decomposition of CPOM. First, we will determine the bacterial and fungal community composition contributing to the decomposition of leaf litter by stable isotope probing and amplicon and metatranscriptome sequencing in the absence of stressors. Thereupon, we will analyse via metatranscriptomics: i) the taxa involved in the decomposition of leaf litter, ii) their specific functional roles, iii) their interactions and redundancies and iv) the effect of multiple stressors on all of the previous aspects.
Subproject A06: "Stressor modulated community responses and functional redundancy of microbial predator-prey interactions"
The subproject A06 focusses on ecophysiological effects of multiple stressors on predator-prey interactions and related shifts of biodiversity, in particular on the effects of multiple stressors on grazing selectivity. The focus will be on protists feeding on bacteria but invertebrates grazing on protists is also adressed. We hypothesize that the stressor-modulated specificity of predator-prey interactions might counter-balance species losses on the community level up to a certain critical level. We investigate i) whether multiple stressors have dominant, additive or interactive effects on biodiversity losses and ii) how critical levels of stressor-induced biodiversity losses affect food selectivity. We will use full factorial replicated time-series experiments to analyse effects between stressor treatments and control treatments during stressor application and after stressor release. In the AquaFlow systems we will analyse community shifts of eukaryotes and bacteria based on amplicon data. Further samples from the different time points will be used for protist food selection experiments using a proteobacterium, an actinobacterium and inert particles. Food selection will be determined using fluorescence in situ hybridization (FISH). Shifts in the food spectrum of macrozoobenthos organisms will be determined based on comparative analyses of gut contents vs. environmental food spectrum using amplicon data. Food selectivity will be calculated using Chessons alpha diversity index both for protists and for macrozoobenthos organisms. Community composition and taxon diversity will be compared in the AquaFlow systems using amplicon sequencing (OTUs as proxy for taxon diversity). Shifts in community composition will be analysed between groups (classes/phyla) and within groups. Community analyses will test for differential effects of stressors on bacterial and eukaryotic microbial communities and further link findings from food selection experiments to community shifts, in particular for the target food organisms, i.e. Actinobacteria and Proteobacteria.
Z02 Maintenance of experimental systems, central field work and central sample analysis
The project will support all experimental and field-based projects of the CRC, by maintaining two central
experimental systems, by performing the general field surveys and by centrally organising and
performing amplicon, metagenomic and metatranscriptomic laboratory analyses.
Within Z02 our working group is responsible for coordinating a series of experiments in the AquaFlow indoor flume system which will be used to test for short-term
stressor increase and release effects under controlled conditions. The AquaFlow system facilitates experiments
with replicated flow channels. Two preparatory experiments will test the variability of results
and the effect of substrate type to set a basis for the data interpretation of the main experiments.
The main experiments will last for nine days; in replicated treatments the effects of current velocity,
temperature increase, salinization and the combination of both stressors on organism groups from
bacteria to benthic invertebrates and their parasites will be tested. Samples of different media will be
taken daily.
The samples will be subjected to a central metagenomics
and metatranscriptomic analysis, and samples from both experiments and the field investigations will
be subject to amplicon analyses. Preparation of amplicon samples will be performed in the Genomic
Core Facility of the Faculty of Biology (UDE), while sequencing will be outsourced. All water samples
from the field studies will be analysed centrally. The Project Z02 will also include central QA/QC components
and closely interact with Z-INF.
Subproject Z-INF: “Data management and integration”
The Z-INF subproject represents the backbone for data management and data integration to facilitate collaboration inside RESIST. Data storage and data exchange between RESIST's individual projects and with external institutions will be coordinated by Z-INF, for which a respective infrastructure will be established. Central data storage facilities will ensure archiving and accessibility of raw and processed data of RESIST. Management of research data for exchange between projects, for publications and future internal and external use will be provided and maintained. For the provided data, data type descriptors with respect to file format, content, sample and experimental affiliation, generation of the data (hardware, software, versions, parameters) will be developed and applied. With mandatory documentation of the generated data, in form of metadata and the introduction of an electronic laboratory notebook connected to the data management system, we will contribute to ensuring reproducible research. In the scope of further utilization of the data, pipelines and tools for data integration will be developed to analyze multiple data sources in conjunction with regard to stressor effects across experimental systems and analyzed organisms. This involves the development of methods to integrate heterogeneous types.
Methods:
- Metabarcoding
- Metatranscriptomics
- FISH
- field sampling
- molecular lab work
- bioinformatic analyses
Project team members:

Differential potential of metabarcoding, metatranscriptomics, and metagenomics for the assessment of lake water quality
The overall objective of this project is to assess the potential of molecular data from microbial communities (metabarcoding, metagenomics and metatranscriptomics) to act as indicators of water quality. This work will be based on an existing dataset comprising samples from 250 European lakes and further field sampling and laboratory work is planned within the project to extend the dataset.
The project entails field sampling, lab work, bioinformatic analyzes of amplicon/metabarcoding, metagenomic and transcriptomic data, as well as statistical analysis of molecular gradients in the data (both in terms of taxonomic and functional diversity), and correlations between these gradients and environmental (physicochemical) variables.
Methods:
- Metabarcoding
- Metatranscriptomics
- Metagenomic
- field sampling
- molecular lab work
- bioinformatic analyses
Project team members:

Auswirkungen von Nanoplastik und Zink auf Mikroorganismen
At the interface between aquatic and anthropogenic areas, precipitation events introduce different substances into bodies of water. This means, among other things, tire abrasion enters waterways. Components such as plastic polymers and zinc are problematic in tire wear. Plastic polymers are broken down into nanoplastics and the zinc concentration increases dramatically. The resulting effect on the environment is currently uncertain.
The preservation of aquatic ecosystems in terms of their biodiversity and ecological functions is essential. Ecological functions are largely carried out by microorganisms and part of the biodiversity comes from this group of organisms. Although microorganisms are among the most common and important organisms, the effects of nanoplastics and zinc have not been sufficiently investigated.
In this project, the consequences of nanoplastics and zinc on microorganisms are examined using next-generation sequencing. On the one hand, changes in the microbial community are examined and on the other hand, changes in gene expressions are examined.
Methods
- Metabarcoding
- Metatranscriptomics
- Laboratory experiments
- Molecular laboratory work
- bioinformatics analyses
Project employees:
The project is part of the NRW Research College-FUTURE WATER
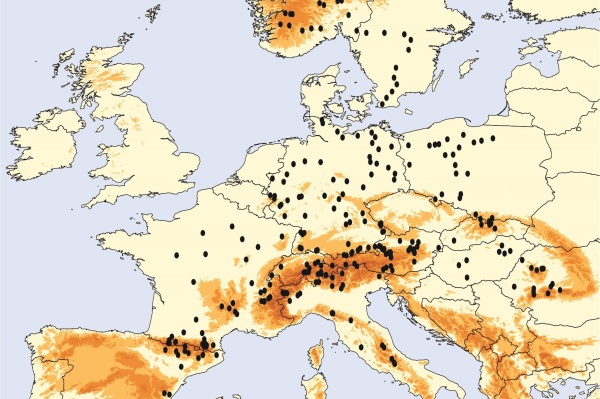
Biogeography of protists
Protists (eukaryotic microbes) fulfill critical ecological functions as they are the dominant primary producers in aquatic environments and, at the same time, are the major consumers of bacteria and thus at the basis of food webs. Further, protist diversity is tremendous but their distribution patterns are not well understood. Doubts about the generalizability of biogeographic patterns as known for animals and plants to other organisms are highly appropriate! To that end, the post-glacial biogeography of Europe is an ideal test case for further testing the generalizability of biogeographic patterns in protists. I this project, we aim to investigate protist distribution patterns in European freshwaters in light of the post-glacial distribution patterns of macro-organisms. We will address variation in protist diversity patterns in natural aquatic ecosystems and relate these changes to environmental factors based on plankton samples from 250 European lakes including lakes from Spain, France, Italy, Switzerland, Austria, Romania, Hungary, the Czech Republic, Slovakia, Poland, Sweden, Norway, Greece, Croatia and Bulgaria. The project will analyze biogeography, phylogeopgraphy and diversity of protist diversity in European freshwater lakes at the community based on massive parallel molecular sequencing data. Taken together the project will test the validity of general biological theories for microbial eukaryotes
Methods
- metabarcoding
- field sampling
- molecular lab work
- bioinformatic analyses
Project employees: