Research
Affinity copolymers for protein surface recognition:
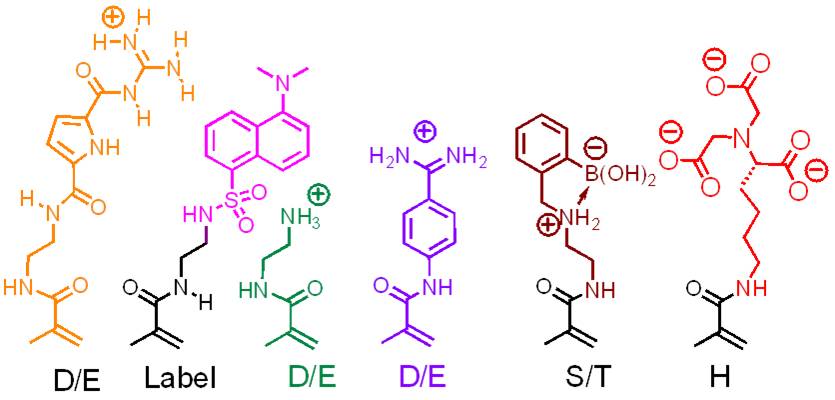
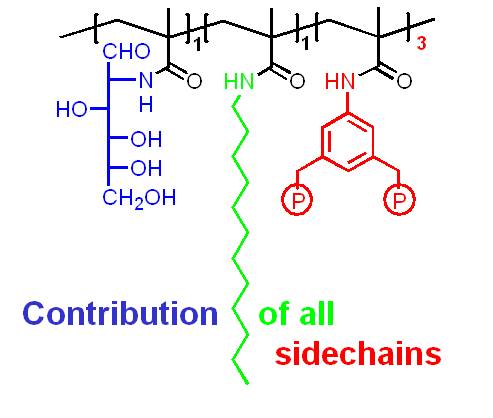
Molecular recognition of larger areas on protein surfaces by artificial binders is a formidable challenge: extended molecular templates must present various binding sites under physiological conditions in such a way, that they can explore the specific arrangement of different amino acid residues both chemically and topologically. A relatively simple approach is the chemical synthesis of a series of monomer building blocks with amino acid selectivity and their copolymerisation in aqueous solution. We have introduced this concept several years ago and have expanded a continuously growing pool of methacrylamides, each of which recognizes an individual class of amino acid residues. Their radical copolymerization in water/DMF mixtures affords linear, water soluble, and fluorescent copolymers in high yields.
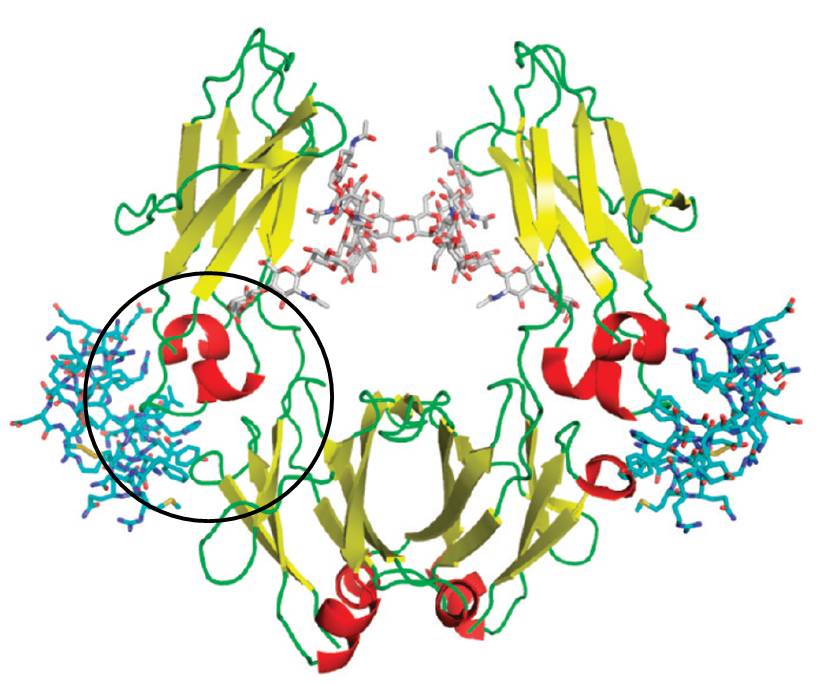
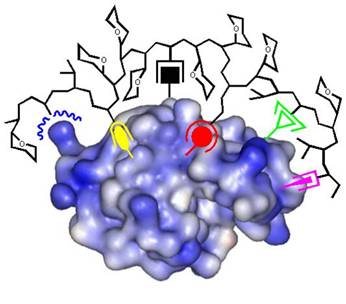
If the appropriate monomer building blocks are mixed at optimal molar ratios, the resulting copolymers are able to bind their target protein with remarkable selectivity and affinity (Lysozyme: Cytochrome c = 100:1; Kd ~ 25 nM). Protein capping can be accompanied by the total shut-down of enzymatic activity, and turned on again by subsquent addition of even more powerful polymer binders.
In close collaboration with the group of H. de Groot we are currently developing selective inhibitors for digestion enzymes to counteract ischemia of the intestines; we also teamed up with M. Ulbricht (UDE) and B. Sellergren (Univ. Dortmund) and designed new affinity polymers as well as capture-phases for medicinal antibodies.
References
- Arginine- and Lysine-Specific Polymers for Protein Recognition and Immobilization. C. Renner, J. Piehler, T. Schrader, J. Am. Chem. Soc. 2006, 128, 620-628.
- Tuning Linear Copolymers into Protein-Specific Hosts. S. Koch, C. Renner, X. Xie, T. Schrader, Angew. Chem. Int. Ed. 2006, 45, 6352-55.
- A Noncovalent Switch for Lysozyme. K. Wenck, T. Schrader, J. Am. Chem. Soc. 2007, 129, 16015-16019.