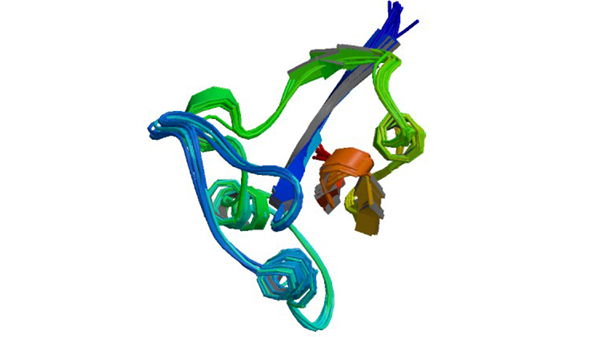
Research
Protein Structure Determination
The department Structural and Medicinal Biochemistry focuses on the elucidation of protein-protein and protein-ligand interactions using biochemical and biophysical strategies in addition to NMR as the main tool for the structure determination of biomolecules. Our research is geared towards predominantly human enzymes/proteins and their macromolecular complexes involved in posttranslational modification processes.

Biochemical Characterisation of Parvulins
Our focus in biochemistry and molecular biology is on parvulin type peptidyl-prolyl cis/trans isomerases (PPIases). The human genome encodes two of these proteins: Pin1 binds to and isomerizes phosphorylated Ser/Thr-Pro motifs and thus influences activity, localization and quality control of phosphorylated proteins. Par14 and its isoform Par17 are implicated in nuclear functions such as chromatin remodeling or RNA processing, in the regulation of receptor-mediated pathways (Par14) as well as in cytoskeleton organization and mitochondrial events (Par17). In addition to these representatives, we are interested in the function and biochemistry of parvulins of unicellular human parasites such as trypanosomes.

Biophysical studies on the interaction of synthetic supramolecular ligands and proteins
In collaboration with research groups from Chemistry, we are currently working on the interaction between proteins and supramolecular ligands such as organic tweezers and branched short polymers. Our interest is to understand and characterize the determinants of binding. With this knowledge, we aim to design proper synthetic ligands suitable for disrupting, stabilizing or modulating protein-protein-interactions.
- Collaborative Research Centre 1093: Supramolecular Chemistry on Proteins (2014 - 2022)
- Collaborative Research Center TRR60: Mutual interaction of chronic viruses with cells of the immune system: from fundamental research to immunotherapy and vaccination (2010 - 2013)
- GRK-1431: Transcription, Chromatin Structure and DNA Repair in Development and Differentiation (2006 - 2016)
- BMBF Project "Structure based design of MRI Probe Molecules for the highly sensitive detection of metastases.” (2009 - 2011)
Entries in the Protein Database (PDB) - NMR
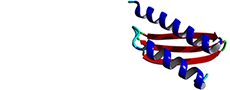
2N84
Solution structure of the FHA domain of TbPar42
Rehic E, Hoenig D, Kamba BE, Goehring A, Hofmann E, Gasper R, Matena A, Bayer P.(2019) Biomolecules 7;9(3).
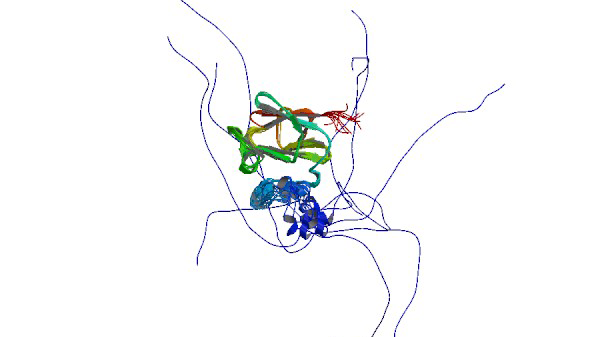
2N87
Solution structure of the PPIase domain of TbPar42
Rehic E, Hoenig D, Kamba BE, Goehring A, Hofmann E, Gasper R, Matena A, Bayer P.(2019) Biomolecules 7;9(3).
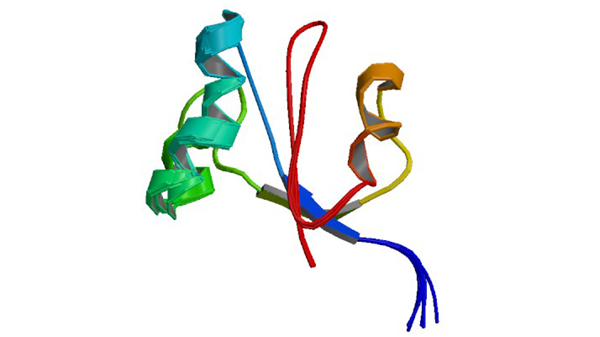
2M08
The solution structure of NmPin, the parvuline of Nitrosopumilus maritimus
Hoppstock L, Trusch F, Lederer C, van West P, Koenneke M, Bayer P. (2016) Bmc Biol. 14: 53-53.
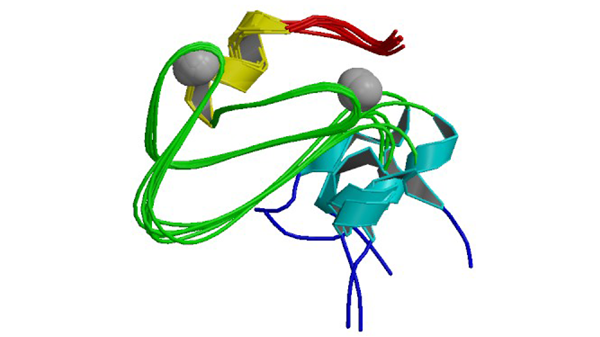
2FNF
C1 domain of Nore1
Harjes E, Harjes S, Wohlgemuth S, Müller KH, Krieger E, Herrmann C, Bayer P. (2006) Structure 14: 881-888.
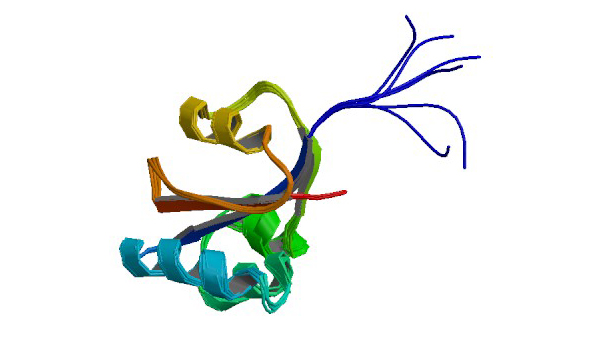
2RQS
3D structure of Pin from the psychrophilic archeon Cenarcheaum symbiosum (CsPin)
Jaremko Ł, Jaremko M, Elfaki I, Mueller JW, Ejchart A, Bayer P, Zhukov I. J. Biol.Chem. 286: 6554-6565.

2K76
Solution structure of a paralog-specific Mena binder by NMR
Link NM, Hunke C, Mueller JW, Eichler J, Bayer P. (2009) Biol.Chem. 390: 417-426.
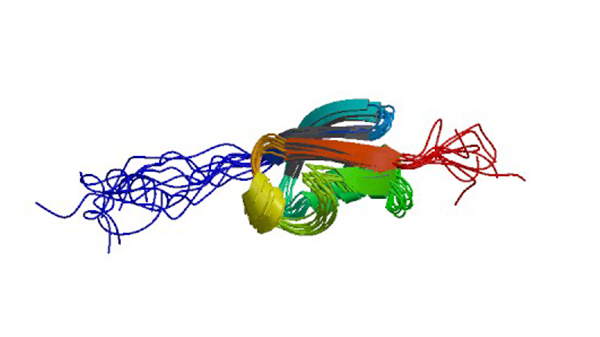
1A5R
Structure determination of the small Ubiquitin-related modifier Sumo-1, NMR, 10 Structures
Bayer P, Arndt A, Metzger S, Mahajan R, Melchior F, Jaenicke R, Becker J. (1998) J. Mol.Biol. 280: 275-286.
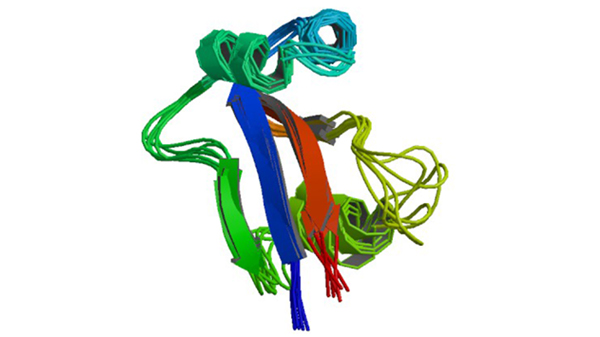
1EQ3
NMR Structure of Human Parvulin HPAR14
Sekerina E, Rahfeld JU, Müller J, Fanghänel J, Rascher C, Fischer G, Bayer P. (2000) J.Mol.Biol. 301: 1003-1017.
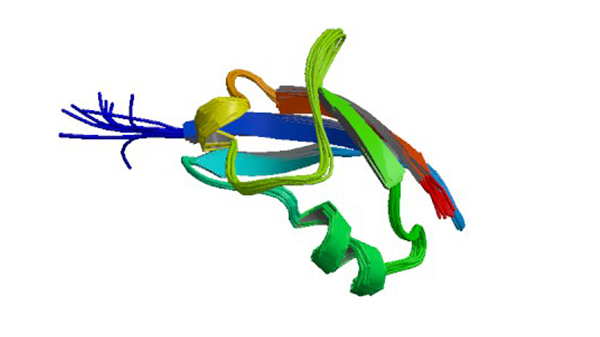
1RLF
Structure determination of the RAS-binding domain of the RAL-specific guanine nucleotide exchange factor RLF
Esser D, Bauer B, Wolthuis RM, Wittinghofer A, Cool RH, Bayer P. (1998) Biochemistry 37: 13453-13462.
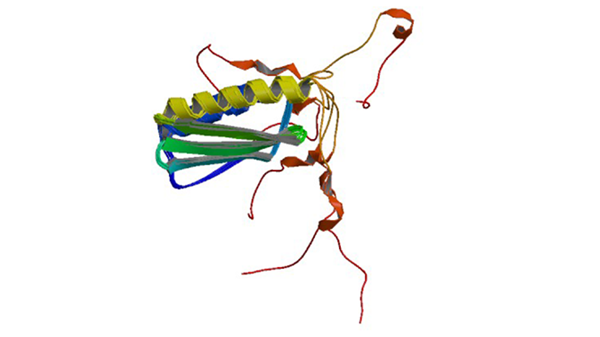
1N3G
Solution structure of the ribosome-associated cold shock response protein Yfia of Escherichia coli
Rak A, Kalinin A, Shcherbakov D, Bayer P. (2002) Biochem. Biophys.Res.Commun. 299: 710-714.
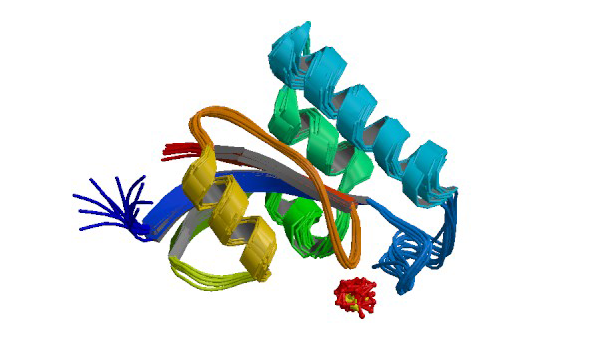
1NMW
Solution structure of the PPIase domain of human Pin1
Bayer E, Goettsch S, Mueller JW, Griewel B, Guiberman E, Mayr LM, Bayer P. (2003) J. Biol. Chem. 278: 26183-26193.
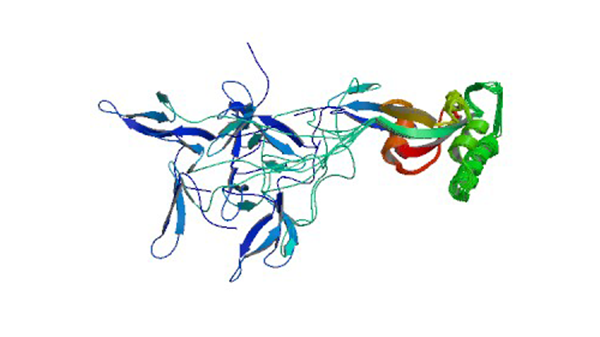
1NMV
Solution structure of human Pin1
Bayer E, Goettsch S, Mueller JW, Griewel B, Guiberman E, Mayr LM, Bayer P. (2003) J. Biol. Chem. 278: 26183-26193.
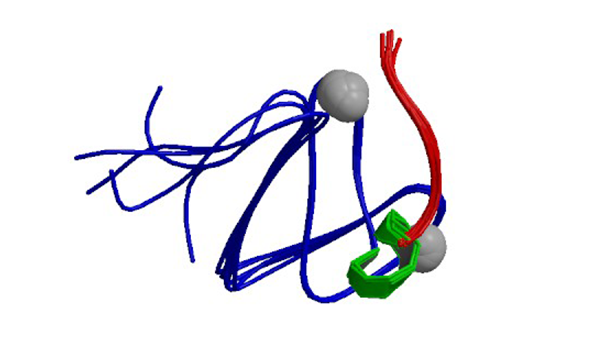
1RFH
Solution structure of the C1 domain of Nore1, a novel Ras effector
Harjes E, Harjes S, Wohlgemuth S, Müller KH, Krieger E, Herrmann C, Bayer P. (2006) Structure. 14(5):881-8.
Entries in the Protein Database (PDB) - X-Ray
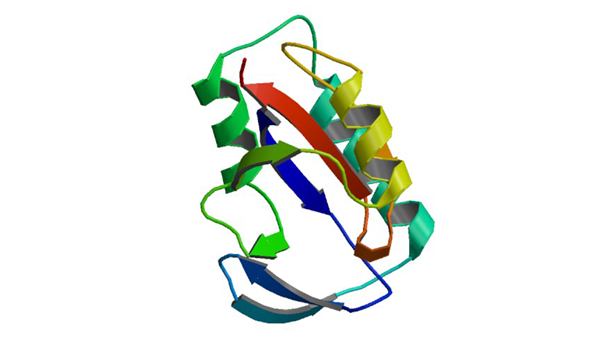
6GMP
Crystal Structure of the PPIASE Domain of TBPAR42
Rehic E, Hoenig D, Kamba BE, Goehring A, Hofmann E, Gasper R, Matena A, Bayer P. Biomolecules. 2019 7;9(3).
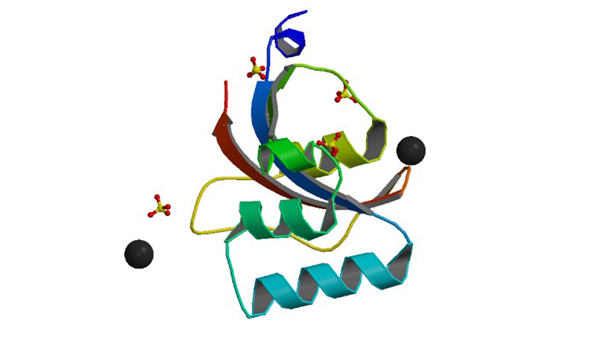
3UI4
0.8 A resolution crystal structure of human Parvulin 14
Mueller JW, Link NM, Matena A, Hoppstock L, Rüppel A, Bayer P, Blankenfeldt W. (2011) J. Am. Chem. Soc. 133: 20096-20099.
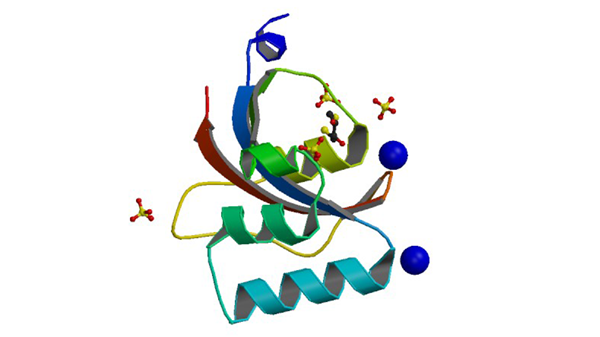
3UI5
Crystal structure of human Parvulin 14
Mueller JW, Link NM, Matena A, Hoppstock L, Rüppel A, Bayer P, Blankenfeldt W. J. Am. Chem. Soc. 133: 20096-20099.
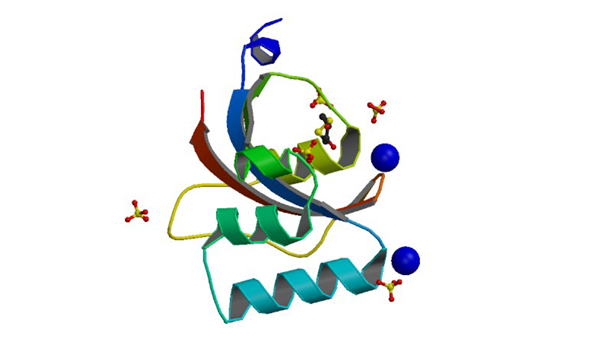
3UI6
0.89 A resolution crystal structure of human Parvulin 14 in complex with oxidized DTT
Mueller JW, Link NM, Matena A, Hoppstock L, Rüppel A, Bayer P, Blankenfeldt W. J. Am. Chem. Soc. 133: 20096-20099.
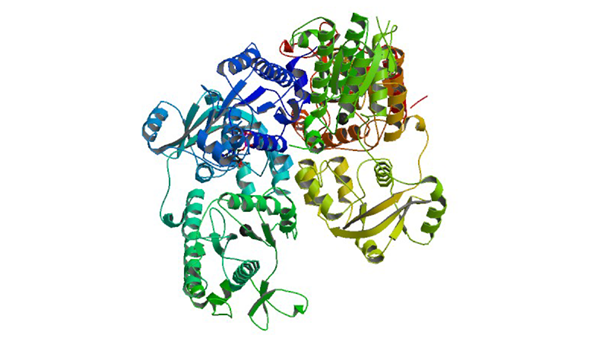
1X6V
The crystal structure of human 3'-phosphoadenosine-5'-phosphosulfate synthetase 1
Harjes S, Bayer P, Scheidig AJ. (2005) J. Mol. Biol. 347: 623-635.
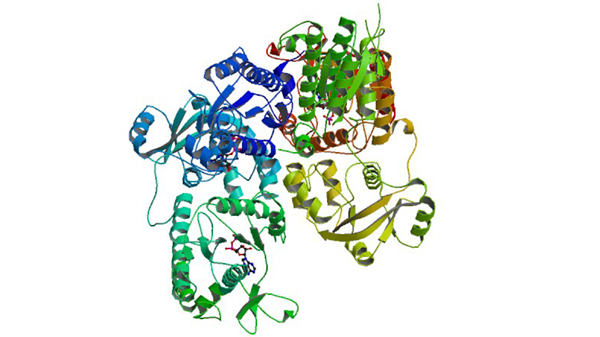
1XJQ
ADP Complex OF HUMAN PAPS SYNTHETASE 1
Harjes S, Bayer P, Scheidig AJ. (2005) J. Mol. Biol. 347: 623-635.
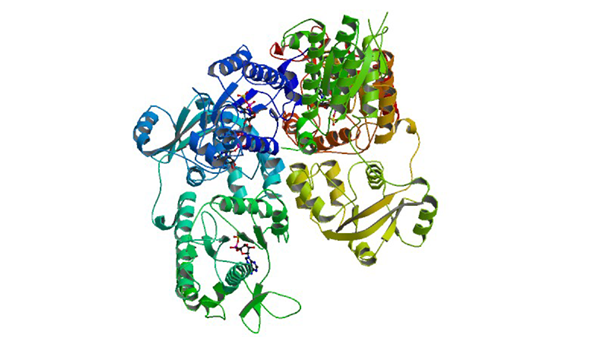
1XNJ
APS complex of human PAPS synthetase 1
Harjes S, Bayer P, Scheidig AJ. (2005) J. Mol. Biol. 347: 623-635.
BMRB Data Bank:
BMRB-11080
BMRB-15946
BMRB-ID 4768
BMRB-ID 4768
BMRB-ID 5979
BMRB-ID 5979
BMRB-ID 6059
BMRB-ID 6059
The network bio-N3MR is a local association of research groups from North Rhine-Westphalia that employ nuclear magnetic resonance (NMR) spectroscopy to study biological macromolecules. The home page of a member group can be reached by clicking on the map.