E. coli Proteases
The E. coli K12 genome encodes at least 74 peptidases/proteases.
Proteases were classified according to their celluar localisation.
View all proteases.
View lists of
Note that some proteases have not yet been classified. Several proteases are not seperately listed because there are only few family members. Also, proteases encoded on plasmids, phages or by pathogenic E. coli are not included in these lists.
All Proteases of E. coli
74 peptidases/proteases are listed. The majority are metalloproteases, followed by Ser proteases, Cys proteases and Asp proteases. Proteases are listed according to their celluar localisation and in alphabetical order. MWs of periplasmic and outer membrane proteases are given for the unprocessed precursor proteins. Clicking on the protein name will open the latest UniProt file.
The topology for each cytoplasmic membrane protein is predicted. Protein sequences are presented in lower case and the TMs in upper case.
Proteases localised in the
Cytoplasm
-
Aminobenzoyl-glutamate utilization protein A (abgA)
Peptidase family M40 InterPro Function Structure MW 46588 Da
436 aa
-
Leucine aminopeptidase LAP (ampA)
Peptidase family M17 InterPro Function Structure MW 54879 Da
503 aa
-
Methionine aminopeptidase (ampM)
Peptidase family M24A InterPro Function Removes the N-terminal methionine from nascent proteins Structure 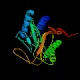 PDB
PDBMW 29331 Da
264 aa
-
Xaa-Pro aminopeptidase/AmpP (pepP)
Peptidase family M24B InterPro Function Releases any N-terminal amino acid preceding proline, even from short peptides Structure 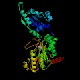 PDB
PDBMW 49684 Da
440 aa
-
ATP-dependent Clp protease proteolytic subunit (clpP)
Peptidase family S14 InterPro Function Heat shock protein, chymotrypsin-like activity, major role in the degradation of misfolded proteins, interacts with chaperone subunits ClpA and X Structure 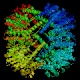 PDB
PDBMW 23186 Da
207 aa
-
Dipeptidyl carboxypeptidase (dcp)
Peptidase family M3 Function Removes dipeptides from the c-termini of oligopeptides, broad specificity, does not hydrolyse bonds in which p1' is Pro, or both p1 and p1' are Gly Structure MW 77384 Da
680 aa
-
D-alanyl-D-alanine dipeptidase (ddpX/vanX)
Peptidase family M45 InterPro Function Hydrolyses Ala-Ala Structure MW 21213 Da
4002 aa
-
ElaD protein (elaD)
Peptidase family SUMO/Ulp1_C InterPro Function Cys protease? Structure MW 45947 Da
402 aa
-
Putative frv operon protein FrvX (frvX)
Peptidase family M42/Glutamyl aminopeptidase (Lactococcus) Function Unclear, homologous to SgcX of ECOLI Structure MW 38733 Da
356 aa
-
Gcp Glycoprotease (gcp/b3064)
Peptidase family M22 InterPro Function Structure MW 36024 Da
356 aa
-
Heat shock protein HslV (hslV)
Peptidase family T1B InterPro Function Thr-Protease subunit of HslUV complex, interacts with chaperone subunit HslU Structure  PDB
PDBMW 18961 Da
175 aa
-
Heat shock protein 31 (hchA / b1967)
Peptidase family Cys protease? InterPro Function Crystal structure reveals a His Asp Cys catalytic triad. Hsp31 was previously characterised as a chaperone. Structure 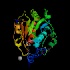 PDB
PDBMW 31059 Da
282 aa
-
Hydrogenase 1 maturation protease (hyaD)
Peptidase family M52 InterPro Function Involved in maturation (C-terminal processing) of HyaB, the subunit of hydrogenase 1 Structure MW 21546 Da
195 aa
-
Hydrogenase 2 maturation protease (hybD)
Peptidase family M52 InterPro Function Involved in maturation (C-terminal processing) of HybB, the subunit of hydrogenase 2 Structure 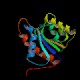 PDB
PDBMW 17751 Da
164 aa
-
Formate hydrogenlyase maturation protein HycH (hycH)
Peptidase family Function Involved in maturation of the large subunit of hydrogenlyase HycE Structure MW 15465 Da
136 aa
-
Hydrogenase 3 maturation protease (hycI)
Peptidase family M52 InterPro Function Involved in maturation of the large subunit of hydrogenlyase HycE Structure MW 16925 Da
155 aa
-
Isoaspartyl dipeptidase IadA (iadA)
Peptidase family M38 Function Hydrolyses l-isoaspartyl (l-beta-aspartyl) dipeptides, to degrade proteins damaged by l-isoaspartyl residue formation Structure 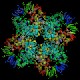 PDB
PDBMW 41084 Da
390 aa
-
Muramoyltetrapeptide carboxypeptidase (LdcA)
Peptidase family unknown Function Converts cytosolic UDP-MurNac-Tetrapeptides to Tripeptides, essential for viability during stationary phase Structure MW 33567 Da
304 aa
-
Lon protease homolog (ycbZ)
Peptidase family s16 Function Lacks ATP ase domain of Lon Structure MW 65818 Da
586 aa
-
Protease Lon (lon)
Peptidase family s16 InterPro Function Heat shock protein, ATP-dependent Ser protease, broad substrate specificity Structure  PDB
PDBMW 87438 Da
784 aa
-
Aminoacyl-histidine dipeptidase/Peptidase D (pepD)
Peptidase family M25 InterPro Function Xaa-His dipeptidase, has specificity for the unusual dipeptide beta-alanyl-l-histidine Structure MW 52784 Da
484 aa
-
Alpha-aspartyl dipeptidase /Peptidase E (pepE)
Peptidase family serine (according to MEROPS) Function Hydrolyses dipeptides containing n-terminal aspartate residues Structure MW 24570 Da
229 aa
-
X-Pro dipeptidase/Peptidase Q (pepQ)
Peptidase family M24B InterPro Function Hydrolysis of Xaa-|-Pro dipeptides, but not on Pro-|-Pro Structure MW 50176 Da
443 aa
-
Aminotripeptidase/Peptidase T (pepT)
Peptidase family M20A InterPro Function Hydrolyzes a tripeptides containing n-terminal Met, Leu, or Phe Structure  PDB
PDBMW 44923 Da
408 aa
-
Hypothetical protease PmbA (pmbA or tldE or b4235)
Peptidase family unknown Function required for the production of the antibiotic peptide MccB17 Structure MW 48369 Da
450 aa
-
Probable zinc protease PqqL (pqqL/b1494)
Peptidase family M16 InterPro Function Structure MW 104656 Da
931 aa
-
Oligopeptidase A (prlC)
Peptidase family M3 InterPro Function Structure MW 77167 Da
680 aa
-
Protease II (ptrB)
Peptidase family s9A InterPro Function Cleaves after Lys and Arg residues Structure MW 79490 Da
686 aa
-
SprT Protein (sprT)
Peptidase family Zn MTpeptdse InterPro Function Structure MW 19348 Da
165 aa
-
Hypothetical protease TldD (tldD or b3244)
Peptidase family unknown Function Structure MW 51364 Da
481 aa
-
Putative metalloprotease YcaL (ycaL)
Peptidase family M48 InterPro Function Homolog of YggG of E. coli Structure MW 26740 Da
254 aa
-
Hypothetical protease YeaZ (yeaZ)
Peptidase family M22 InterPro Function Structure MW 25181 Da
231 aa
-
Putative protease YegQ (yegQ)
Peptidase family U32 InterPro Function Structure MW 51193 Da
453 aa
-
Hypothetical protein YgeY (ygeY)
Peptidase family M20A InterPro Function Structure MW 44804 Da
403 aa
-
Putative metalloprotease YggG (yggG)
Peptidase family M48 InterPro Function Homolog of YcaL of E. coli Structure MW 26842 Da
252 aa
-
Putative Cys protease YhbO (yhbO)
Peptidase family c40 Function Structure MW 18858 Da
172 aa
-
Hypothetical protein YibP (yibG)
Peptidase family M37 InterPro Function Structure MW 46594 Da
419 aa
-
Hypothetical protein YpdF (ypdF)
Peptidase family M24 InterPro Function Structure MW 39624 Da
361 aa
Cytoplasmic membrane
-
Protease DegS (degS)
Peptidase family S2C InterPro Function Essential serine protease, Cleaves RseA to induce sigma E cascade Structure  PDB
PDB
transmembrane domains: 1 (N-in)
TMHMM gives 5-27
SP gives signal seq.
mfvkllrsVA IGLIVGAILL VAMPSLrsln plstpqfdst detpasynla vrraapavvn
vynrglntns hnqleirtlg sgvimdqrgy iitnkhvind adqiivalqd grvfeallvg
sdsltdlavl kinatgglpt ipinarrvph igdvvlaign pynlgqtitq giisatgrig
lnptgrqnfl qtdasinhgn sggalvnslg elmgintlsf dksndgetpe gigfaipfql
atkimdklir dgrvirgyig iggreiaplh aqgggidqlq givvnevspd gpaanagiqv
ndliisvdnk paisaletmd qvaeirpgsv ipvvvmrddk qltlqvtiqe ypatnMW 37581 Da
355 aa
-
FtsH (ftsH/hflB)
Peptidase family M41/AAA Protease InterPro Function essential ATP-dependent zinc metalloprotease, cleaves soluble and integral membrane proteins Structure 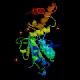 PDB
PDB
transmembrane domains:2 (potential)TMHMM gives 5-24 97-119
SP gives 5-24 96-120maknLILWLV IAVVLMSVFQ SFgpsesngr kvdystflqe vnndqvrear
ingreinvtk kdsnryttyi pvqdpklldn lltknvkvvg eppeepSLLA
SIFISWFPML LLIGVWIFFm rqmqggggkg amsfgkskar mltedqiktt
fadvagcdea keevaelvey lrepsrfqkl ggkipkgvlm vgppgtgktl
200
lakaiageak vpfftisgsd fvemfvgvga srvrdmfeqa kkaapciifi
deidavgrqr gaglggghde reqtlnqmlv emdgfegneg iiviaatnrp
dvldpallrp grfdrqvvvg lpdvrgreqi lkvhmrrvpl apdidaaiia
rgtpgfsgad lanlvneaal faargnkrvv smvefekakd kimmgaerrs
400
mvmteaqkes tayheaghai igrlvpehdp vhkvtiiprg ralgvtfflp
egdaisasrq klesqistly ggrlaeeiiy gpehvstgas ndikvatnla
rnmvtqwgfs eklgpllyae eegevflgrs vakakhmsde tariidqevk
aliernynra rqlltdnmdi lhamkdalmk yetidapqid dlmarrdvrp
600
pagweepgas nnsgdngspk aprpvdeprt pnpgntmseq lgdkMW 70708 Da
644 aa
-
GlpG (glpG)
Peptidase family s54 InterPro Function Structure transmembrane domains: 6 (potential)
Phobius gives 96-115 135-160 172-188 194-212 224-242 248-267
SP gives none
mlmitsfanp rvaqafvdym atqgviltiq qhnqsdvwla desqaervra
Elarflenpa dprylaaswq aghtgsglhy rrypffaalr eragpVTWVM
MIACVVVFIA MQILGdqevm lwlawpfdpt lkfeFWRYFT HALMHFSLMH
ILFNLLWWWY LGGAVekrlg sgkLIVITLI SALLSGYVQQ kfsgpWFGG
SGVVYALMGY VWLrgerdpq sgIYLQRGLI IFALIWIVAG WFdlfGMSMA
NGAHIAGLAV GLAMAFVdsl narkrkMW 31307 Da
276 aa
-
Heat shock protein HtpX (htpX)
Peptidase family M48 InterPro Function Structure transmembrane domains: 4 (potential)
TMHMM gives 5-27 32-54 158-180 195-217
SP gives 7-27 35-51 158-178 193-213
mmriALFLLT NLAVMVVFGL VLSLTGIqss sVQGLMIMAL LFGFGGSFVS
LLMSkwmalr svggevieqp rnererwlvn tvatqarqag iampqvaiyh
apdinafatg arrdaslvav stgllqnmsp deaeaviahe ishiangdmv
tmtliqgVVN TFVIFISRIL AQLAAGFMGG nrdegeesng npliYFAVAT
200
VLELVFGILA SIITMWFsrh refhadagsa klvgrekmia alqrlktsye
pqeatsmmal cingksksls elfmthppld kriealrtge ylkMW 31923 Da
293 aa
-
Leader peptidase Lep4/Prepilin-like protein (hofD/hopD)
Peptidase family A24 InterPro Function Leader peptidase, cleaves type-4 fimbrial leader sequence Structure transmembrane domains: 7 (potential), SP has 6
TMHMM gives 7-29 49-71 78-100 104-122 127-149 169-191 204-223
SP gives 3-23 68-88 104-124 128-148 175-195 203-223
mtMLLPLFIL VGFIADYFVN AIAYhlsple dktaltfrqv lvhfrqkkYA
WHDTVPLILC VAAAIACALA PFtpIVTGAL FLYFCFVLTL SVIdfrtqll
pdkLTLPLLW LGLVFNAQYG LIdlhdaVYG AVAGYGVLWC VYWGVWLVCh
keglgygdfk llaaagaWCG WQTLPMILLI ASLGGIGYAI Vsqllqrrti
200
ttIAFGPWLA LGSMINLGYL AWIsyMW 24957 Da
225 aa
-
Leader peptidase I (LepB)
Peptidase family s26 InterPro Function General leader peptidase Structure  PDB
PDB
transmembrane domains: 2 (N-out)
TMHMM gives 5-27 60-82
SP gives 4-22 59-77
manmFALILV IATLVTGILW CVDKFFFapk rrerqaaaqa aagdsldkat
lkkvapkpgW LETGASVFPV LAIVLIVRSF IYepfqipsg smmptlligd
filvekfayg ikdpiyqktl ietghpkrgd ivvfkypedp kldyikravg
lpgdkvtydp vskeltiqpg cssgqacena lpvtysnvep sdfvqtfsrr
200
nggeatsgff evpknetken girlserket lgdvthrilt vpiaqdqvgm
yyqqpgqqla twivppgqyf mmgdnrdnsa dsrywgfvpe anlvgratai
wmsfdkqege wptglrlsri ggihMW 35960 Da
324 aa
-
Lipoprotein signa l peptidase (lspA)
Peptidase family A8 InterPro Function Leader peptidase, removes signal peptides from lipoproteins Structure transmemebrane domains: 4 (potential)
TMHMM gives 13-35 67-86 99-121 136-158
SP gives 12-26 70-88 96-113 142-159
msqsicstgl rwLWLVVVVL IIDLGSKYLI LQNFAlgdtv plfpslnlhy
arnygaafsf ladsggWQRW FFAGIAIGIS VILAVMMYrs katqklnnIA
YALIIGGALG NLFDRLWHGF Vvdmidfyvg dwhfaTFNLA DTAICVGAAL
IVLEGFLPsr akkqMW 18156 Da
164 aa
-
Leader peptidase PppA (pppA)
Peptidase family A24 InterPro Function Leader peptidase, removes signal peptides from lipoproteins Structure transmemebrane domains: 7 (potential)
TMHMM gives 15-37 102-120 124-141 148-167 177-196 209-231 246-268
SP gives 0
mlfdvfqqyp tampVLATVG GLIIGSFLNV VIWrypimlr qqmaefhgem
ssaqskisla lprshcphcq qtirirdnip lfswlmlkgr crdcqakisk
rYPLVELLTA LAFLLASLVW pesgWGLAVM ILSAWLIAAS VIdldhqWLP
DVFTQGVLWT GLIAAWAqqs pltlqdAVTG VLVGFITFYS LRWIAGIVLr
200
kealgmgdVL LFAALGGWVG ALSLPNVALI Asccgliyav itkrgstTLP
FGPCLSLGGI ATLYLQALFMW 29466 Da
269 aa
-
Possible Ser protease SohB (sohB)
Peptidase family s49 InterPro Function Suppressor of degP(htrA) null mutation Structure transmembrane domains: 1 (potential), TMHMM has 2
TMHMM gives 10-32 190-207
SP gives 9-29
mellseyglF LAKIVTVVLA IAAIAAIIVN VAqrnkrqrg elrvnnlseq
ykemkeelaa almdshqqkq whkaqkkkhk qeakaakaka klgevatdsk
prvwvldfkg smdahevnsl reeitavlaa fkpqdqvvlr lespggmvhg
yglaasqlqr lrdknipltv tvdkvaasgg ymmacvadki vsapfaivgs
200
igvvaqmpnf nrflkskdid ielhtagqyk rtltllgent eegrekfree
lnethqlfkd fvkrmrpsld ieqvatgehw ygqqavekgl vdeintsdev
ilslmegrev vnvrymqrkr lidrftgsaa esadrlllrw wqrgqkplmMW 39366 Da
349 aa
-
Signal peptide peptidase/Protease IV (sppA)
Peptidase family s49 InterPro Function Degrades cleaved signal peptides Structure transmembrane domains: 3 (potential), TMHMM has 1
TMHMM gives 21-43
SP gives 29-45 398-414 421-441
mrtlwrfiag ffkwtwrlln FVREMVLNLF FIFLVLVGVG IWMqvsggds
ketasrgall ldisgvivdk pdssqrfskl srqllgassd rlqenslfdi
vntirqakdd rnitgivmdl knfaggdqps mqyigkalke frdsgkpvya
vgenysqgqy ylasfankiw lspqgvvdlh gfatnglyyk slldklkvst
200
hvfrvgtyks avepfirddm spaareadsr wigelwqnyl ntvaanrqip
aeqvfpgaqg llegltktgg dtakyalenk lvdalassae iekaltkefg
wsktdknyra isyydyalkt padtgdsigv vfangaimdg eetqgnvggd
ttaaqirdar ldpkvkaivl rvnspggtvt aseviraela aaraagkpVV
400
VSMGGMAASG GYWIstpanY IVANPSTLTG SIGIFGVITT Vensldsigv
htdgvstspl advsitralp peaqlmmqls iengykrfit lvadarhstp
eqidkiaqgh vwtgqdakan glvdslgdfd davakaaela kvkqwhleyy
vdeptffdkv mdnmsgsvra mlpdafqaml paplasvast vksesdklaa
600
fndpqnryaf cltcanmrMW 67233 Da
618 aa
-
rseP (YaeL)
Peptidase family M50B Function RIP protease, degrades the antisigma factor RseA Structure transmembrane domains: 4 (potential), SP has 3
TMHMM gives 2-21 98-120 376-398 426-445
SP gives 107-127 376-396 430-450
mLSFLWDLAS FIVALGVLIT Vhefghfwva rrcgvrverf sigfgkalwr
rtdklgteyv ialiplggyv kmlderaepv vpelrhhafn nksvgqrAAI
IAAGPVANFI FAIFAYWLVF iigvpgvrpv vgeiaansia aeaqiapgte
lkavdgietp dwdavrlqlv dkigdestti tvapfgsdqr rdvkldlrhw
200
afepdkedpv sslgirprgp qiepvlenvq pnsaaskagl qagdrivkvd
gqpltqwvtf vmlvrdnpgk slaleierqg splsltlipe skpgngkaig
fvgiepkvip lpdeykvvrq ygpfnaivea tdktwqlmkl tvsmlgklit
gdvklnnlsg pisiakgagm taelgVVYYL PFLALISVNL GIINLFPLpv
400
ldgghllfla iekikggpvs ervqdFCYRI GSILLVLLMG LALFNdfsrlMW 49071 Da
450 aa
-
YfbL (yfbL)
Peptidase family M28A Function Structure transmembrane domains: 1 (potential)
TMHMM gives 5-27
SP gives 4-24
mkkIIFAFII LFVFLLPMII FYQPWVnalp stprhaspeq lektvryltq
tvhprsadni dnlnrsaeyi kevfvssgar vtsqdvpitg gpyknivady
gpadgpliii gahydsassy endqltytpg addnasgvag llelarllhq
qvpktgvqlv ayaseeppff rsdemgsavh aaslerpvkl mialemigyy
200
dsapgsqnyp ypamswlypd rgdfiavvgr iqdinavrqv kaallssqdl
svysmntpgf ipgidfsdhl nywqhdipai mitdtafyrn kqyhlpgdta
drlnyqkmaq vvdgvitlly nskMW 35931 Da
323 aa
Periplasm
-
PBP-5, D-alanyl-D-alanine carboxypeptidase fraction A (dacA)
Peptidase family s11 Function Cell wall formation Structure 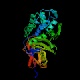 PDB
PDBMW 44444 Da
403 aa (1-29 signal pept.)
-
PBP-4, D-alanyl-D-alanine carboxypeptidase (dacB)
Peptidase family s13 Function RIP protease, degrades the antisigma factor RseA Structure MW 51798 Da
477 aa (1-20 signal pept.)
-
PBP-6, D-alanyl-D-alanine carboxypeptidase fraction C (dacC)
Peptidase family s11 Function Cell wall formation Structure MW 43609 Da
400 aa (1-27 signal pept.)
-
PBP-6B, D-alanyl-D-alanine carboxypeptidase (dacD )
Peptidase family s11 Function Cell wall formation Structure MW 43346 Da
388 aa (1-21 signal pept.)
-
Protease Do (degP/htrA)
Peptidase family S2C Function Heat shock protein, serine protease and chaperone ectivities, required at high temperature, virulence factor, involved in refolding and degradation of damaged proteins Structure  PDB
PDBMW 49439 Da
474 aa (1-26 signal pept.)
-
Protease DegQ (degQ)
Peptidase family s2c Function Serine protease Structure 2 PDZ domains MW 47205 Da
455 aa (1-27 signal pept.)
-
Alkaline phosphatase isozyme conversion protein IAP (iap or b2753)
Peptidase family M28C Function Structure MW 37920 Da
345 aa (1-24 signal pept.)
-
Penicillin-insensitive D-alanyl-D-alanine-endopeptidase (mepA)
Peptidase family Unknown Function Cell wall formation Structure MW 30136 Da
274 aa (1-19 signal pept.)
-
Putative Cys protease NlpC (nlpC)
Peptidase family Unknown Function Structure potential lipoprotein MW 17283 Da
154 aa (1-15 signal pept.)
-
PBP-7, D-alanyl-D-alanine-endopeptidase (pbpG or b2134)
Peptidase family Unknown Function Cell wall formation Structure MW 34245 Da
313 aa (1-28 signal pept.)
-
Tail-specific protease Prc/Tsp (tsp)
Peptidase family s41 Function Degrades proteins containing a 11 residue C-term. ssrA degradation tag Structure 1 PDZ domain MW 76663 Da
682 aa (1-22 signal pept.)
-
Protease III (ptrA)
Peptidase family M16 Function Endopeptidase, may have preference for small peptides Structure MW 107708 Da
962 aa (1-23 signal pept.)
-
Protease I (tesA)
Peptidase family Lipase Function Hydrolyzes only long chain acyl thioesters (C12-C18), similar to chymotrypsin Structure 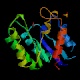 PDB
PDBMW 23622 Da
208 aa (1-26 signal pept.)
-
UmuD (umuD)
Peptidase family S24 Function Structure 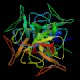 PDB
PDBMW 15063 Da
139 aa (1-24 signal pept.)
-
Putative Cys protease YafL (yafL)
Peptidase family C40 Function Structure potential lipoprotein MW 28785 Da
249 aa (1-17 signal pept.)
-
Putative protease YdcP (ydcP)
Peptidase family U32 Function Structure MW 72701 Da
653 aa (1-20 signal pept.)
-
Putative protease YdgD (ydgD)
Peptidase family S2B Function Structure MW 29277 Da
273 aa (1-21 signal pept.)
-
Putative Cys protease YdhO (ydhO)
Peptidase family C40 Function Structure MW 29916 Da
271 aa (1-27 signal pept.)
-
Hypothetical protein YebA (yebA)
Peptidase family M37 Function Structure MW 49057 Da
440 aa (1-34 signal pept.)
-
Putative protease YhbU (yhbU)
Peptidase family U32 Function Structure MW 37047 Da
331 aa (1-15 signal pept.)
-
Protein YhjJ (yhjJ)
Peptidase family M16 Function Structure MW 55527 Da
498 aa (1-24 signal pept.)
Outer membrane
-
Lipoprotein NlpD (nlpD)
Peptidase family M37 Function Structure MW 40149 Da
379 aa (1-25 signal pept.)
-
OmpT/ProteaseVII (ompT)
Peptidase family S18 Function The 3D structure questions that OmpT is a ser protease. Structure  PDB
PDBMW 35562 Da
317 aa (1-20 signal pept.)
