Teaching
Teaching
Biophysics and AI tools in Biology
The course is structured around two main areas:
Biophysics: This section delves into the fundamental physical principles that govern living systems, providing an introduction to equilibrium thermodynamics and statistical mechanics. These concepts will be utilized to gain both qualitative and semi-quantitative insights into biomolecules, employing a two-state model for analysis.
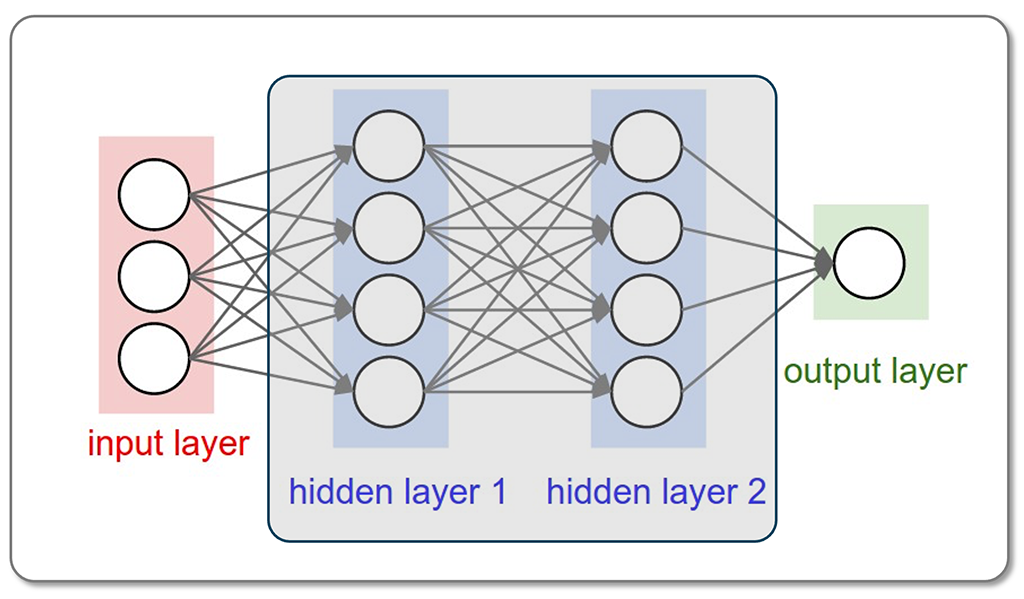
Artificial Intelligence in Biology: This part introduces AI tools, particularly Machine Learning and Deep Learning, that are essential for analyzing large biological datasets. Students learn how to use the AlphaFold model for predicting protein structures and their complexes.
In addition to the core content, the course includes interactive exercises and online materials, promoting collaborative analysis and discussion of exercise solutions among participants.
DNA Nanotechnology
(Practical course)
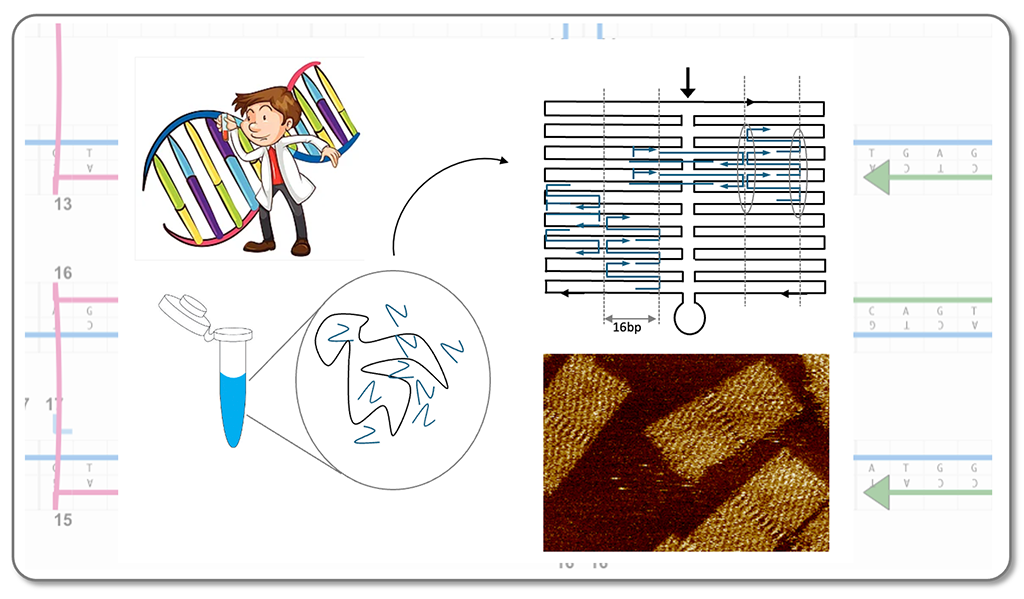
The course offers an introduction to DNA nanotechnology, covering its historical background and the key design strategies, including multi-stranded approaches and scaffolded-DNA origami. The students learn to design simple DNA motifs and planar DNA origami structures and modify these structures to create specific protein patterns. In the lab, students learn various purification methods for DNA origami and explore how experimental conditions affect structure quality and yield. Analysis of the structures is done using agarose-gel electrophoresis and atomic force microscopy. The course also covers toehold-mediated strand displacement and its application in conformational changes monitored by FRET spectroscopy, as well as single-molecule force experiments to stretch DNA until rupture. The course ends with seminars based on current literature in the field.
Single Molecule Techniques
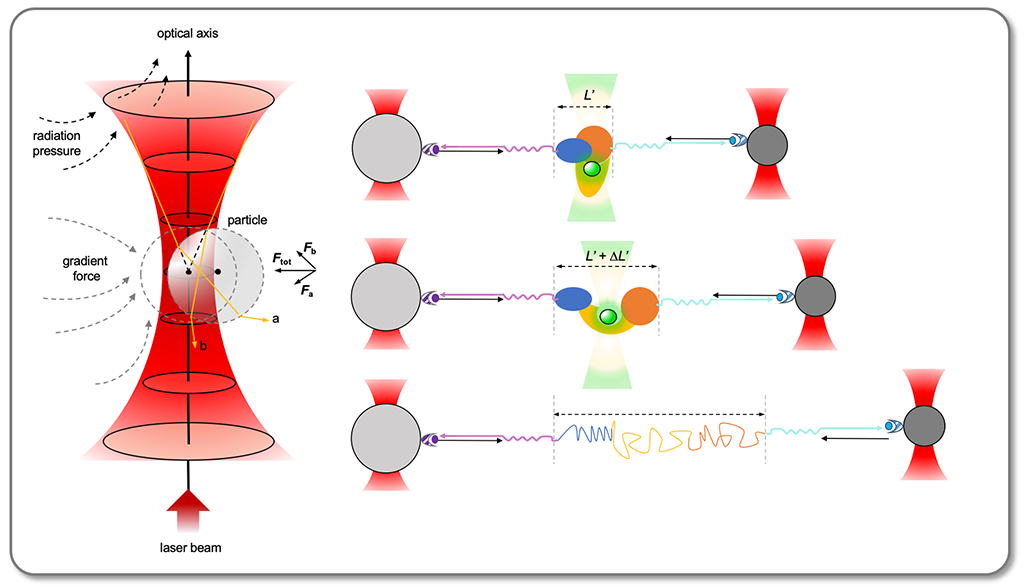
This course provides an introduction to modern single-molecule techniques, focusing on force- and optical-based methods like atomic force microscopy, optical/magnetic tweezers, and super-resolution techniques (STED, STORM, PALM). The course explains the physical principles behind these technologies and their applications in biology, aiming to connect individual molecular behavior to larger interactions involving proteins, small molecules, and DNA, as well as studying conformational changes and molecular motor dynamics.
The course also includes seminars on current literature, where students conduct literature searches, present findings, and engage in discussions to deepen their understanding of single-molecule techniques and their biological implications.
Mathematical Models in Molecular Biology
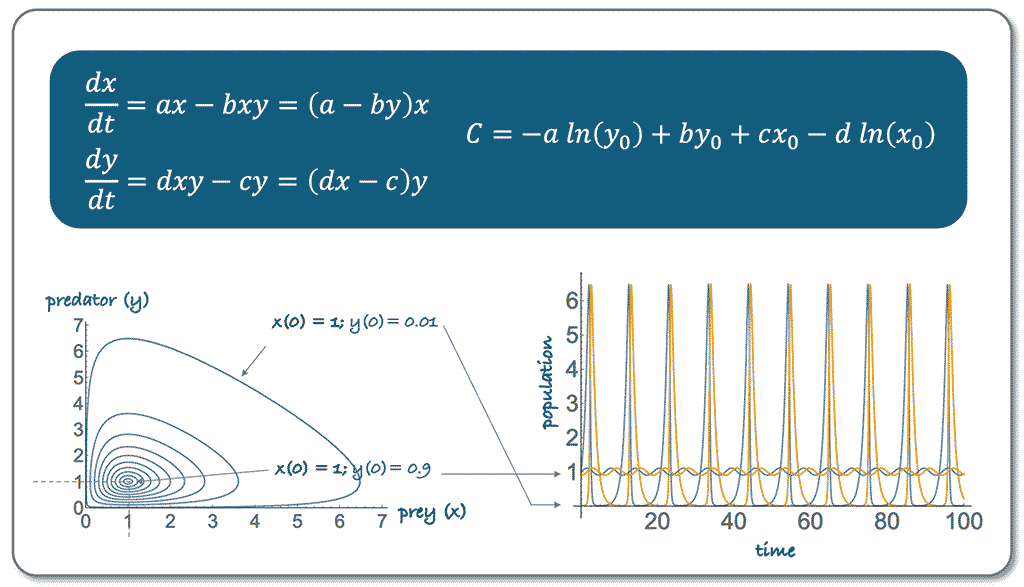
The course introduces students to basic mathematical models used in modern biology, covering topics such as population dynamics, biochemical reactions, enzyme kinetics, and biological oscillators. Students learn about models like exponential growth, logistic growth, predator-prey interactions, and key concepts such as steady state and bifurcation. The accompanying exercise session provides practical applications of these models through numerical examples and simulations, helping students visualize the effects of varying parameters on biological scenarios.
Molecular and Cellular Biophysics
The course provides students with a physical perspective on biological phenomena by introducing fundamental principles that govern living systems. It covers key concepts such as the elastic properties of biopolymers, the regulation of DNA transcription, and viral packing through models like the spring model and the random walk. Additionally, it explores translational and rotary molecular motors, ion channels, and cellular machineries in relation to equilibrium thermodynamics and statistical mechanics. The course concludes with an overview of biological pattern formation and cellular differentiation. It incorporates state-of-the-art biophysical techniques, including atomic force microscopy and single-molecule fluorescence microscopy, along with interactive exercises and online resources.