Research
Current Directions
Nanobodies & Targeted Protein Degradation
Our research focuses on developing nanobody-based strategies and targeted protein degradation (TPD) technologies to understand and manipulate cellular processes with high precision. Nanobodies, derived from camelid heavy-chain antibodies, are small, stable, and highly specific binders that can penetrate cells and access compartments inaccessible to conventional antibodies. We exploit these properties to generate nanobodies that act as inhibitors, biosensors, or recruiters of degradation machinery, thereby allowing the selective modulation or removal of proteins directly at their site of action.
By integrating nanobody engineering with innovative delivery systems, protein localization signals, and chemical biology approaches, we aim to establish new strategies for both reversible protein interference and acute protein depletion. In this context, we also develop and apply proteolysis-targeting chimeras (PROTACs), which redirect the cell’s degradation machinery toward selected targets. These tools provide unique opportunities to study protein function in real time and to dissect dynamic signaling pathways, while also offering translational potential for targets traditionally considered “undruggable.”
Our methodological framework combines advanced live-cell imaging with phenotypic, functional, and biochemical protein–protein interaction assays. Complemented by nanobody-based probes and switchable chemical modulators, this interdisciplinary approach allows us to investigate protein function across different spatial and temporal scales. Looking ahead, we see nanobodies and TPD technologies as central elements of next-generation precision therapies. By bridging fundamental cell biology with translational applications, our work aims to provide new avenues for highly specific interventions in oncology and other biomedical fields.
Survivin
Due to its dual role as an apoptosis inhibitor and mitotic regulator, Survivin is accepted as a nodal protein hub, with high relevance in basic cell and translational cancer biology. Particularly, our work on the impact of Survivin's nuclear export signal not only resulted in several publications but also provided the basis for innovative chemico-genetic translational interference strategies. Currently, we are pursuing the concept of supramolecular inhibition by rational chemical design and specific nanobodies combined with innovative nanocarrier based delivery tools. In addition, we recently revealed a so far unknown role in the DNA damage response and as such now set the stage for in-depth investigations of Survivin's role in cancer biology and therapy response. We additionally focus on Survivin’s apoptotic/mitotic switch, whether it really exists as proposed and how it can be induced. This has so far eluded experimental investigation, although it would be of utmost importance to finally understand how Survivin decides between its different biological roles.
Taspase1
Although less is known about the molecular (patho)mechanisms of the threonine protease Taspase1, the ZMB’s outstanding expertise in protease research represents an ideal platform to study its unresolved impact on ‘Cell fate decisions in development and disease’. Employing cutting-edge live cell microscopy (in cooperation with our high-end microscopes implemented at the Imaging Center Campus Essen) and biosensor systems, we not only uncovered novel mechanisms regulating Taspase1’s structure activity relationships (SAR), but also identified previously unknown targets with cellular functions in e.g., transcriptional regulation and cell differentiation and migration. Hence, Taspase1 also represents an important molecular switch determining cell fate and in addition serves as a valuable model in protease research to unravel novel fine-tuning mechanisms with translational relevance.
Currently, we are pursuing the concept of supramolecular inhibition of pivotal Taspase1 protein interactions. Here, we particularly focus on its Importin-mediated nuclear transport, but also on the interplay of prerequisites for autocatalytic activation of the protease and proteolytic activity in general. Is it possible to interfere with Importin-related nucleolar association and at which steps of the protease activation process? And finally, how is substrate specificity for only one or a certain subset of Taspase1 target(s) achieved and at which developmental stages? Therefore, we are engaging in developing novel nanobodies targeting Taspase1 to allow specific modulation as well as timely induction of targeted protein degradation (TPD).
Recently, we succeeded in the development of supramolecular binders blocking the pivotal interaction of Taspase1's nuclear import signal (NLS) with the import receptor Importin α ( see below).
Published Results
Nanobodies
- Small but mighty: the versatility of nanobodies in modern medicine
- Nanobodies – effektive Immunwaffen gegen Krebs
Survivin
- Chromosomal passenger proteins engage in translesion synthesis
- Tuning Nanobodies’ bioactivity against Survivin
Taspase1
Small but mighty: the versatility of nanobodies in modern medicine
Nanobodies, the single-domain fragments derived from camelid antibodies, are increasingly recognized as a transformative class of biomolecules. Their compact size, exceptional stability, and ability to access otherwise hidden epitopes provide clear advantages over conventional antibodies. These properties have enabled applications ranging from molecular imaging and diagnostics to targeted drug delivery and intracellular protein modulation. After decades of development, nanobody-based therapeutics are now moving into the clinic, with the approval of caplacizumab marking an important milestone in their medical relevance.
This review highlights the structural and functional features that make nanobodies uniquely versatile, alongside recent engineering strategies that expand their scope. Advances such as bispecific formats, nanobody–drug conjugates, and intracellularly active variants are opening new opportunities in oncology, neurodegenerative, infectious, and autoimmune diseases. Looking ahead, integration with cutting-edge modalities including targeted protein degradation, CAR-T cell therapy, and gene editing points to their potential to address previously “undruggable” targets. Although challenges remain in delivery, manufacturing, and regulatory approval, rapid progress in AI-driven design, high-throughput display technologies, and smart delivery systems is paving the way for next-generation nanobody therapeutics. Together, these developments position nanobodies as powerful tools at the forefront of precision medicine and future biologics.
Nanobodies – effektive Immunwaffen gegen Krebs
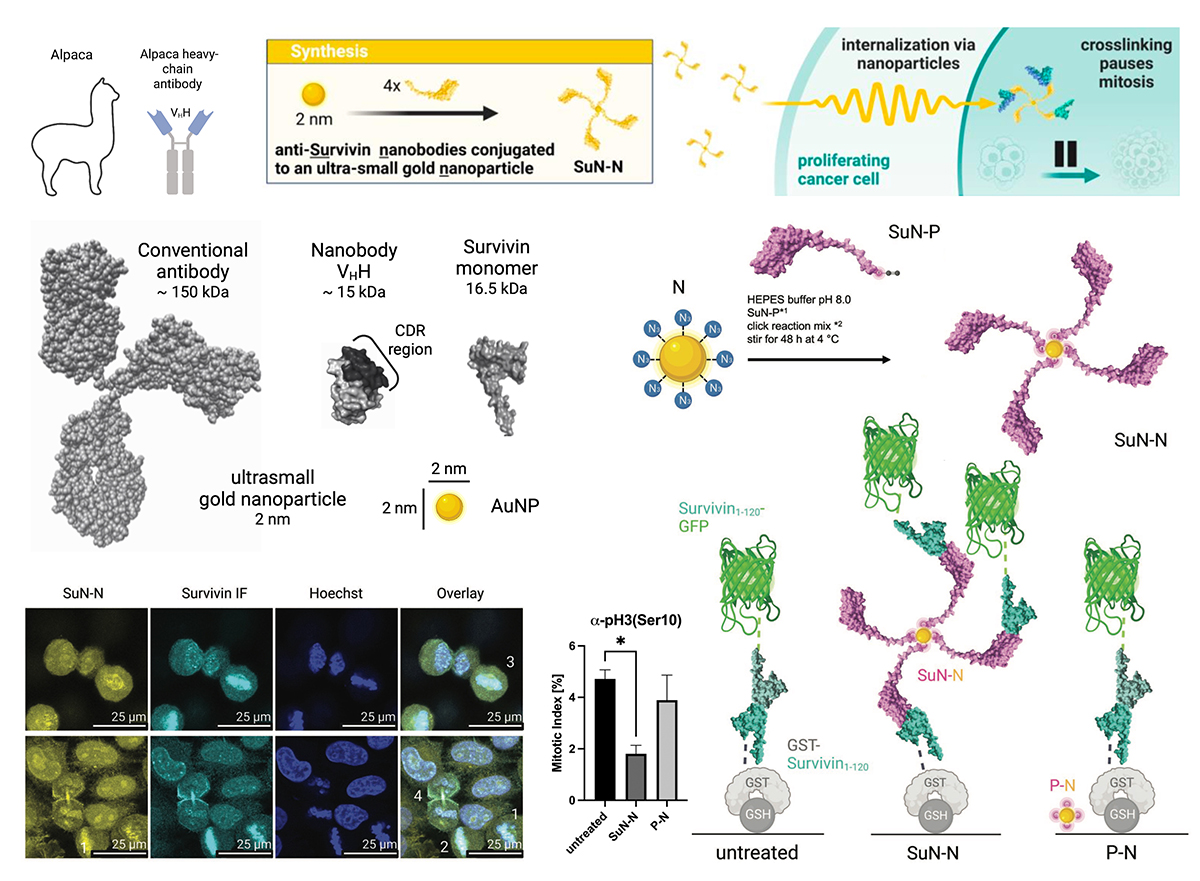
Recently, we attached a nanobody targeting the cancer-relevant protein Survivin to ultrasmall gold nanoparticles. This enabled intracellular uptake, Survivin crosslinking, and interference with cancer cell mitosis. Coupling of nanobodies to nanosized scaffolds is universally applicable to improve the function and therapeutic potential of nanobodies in general. Currently, we are developing novel inhibitory nanobodies against Survivin as well as the oncologically relevant Protease Taspase 1.
Interview mit Prof. Dr. Shirley Knauer auf BIOspektrum
Chromosomal passenger proteins engage in translesion synthesis
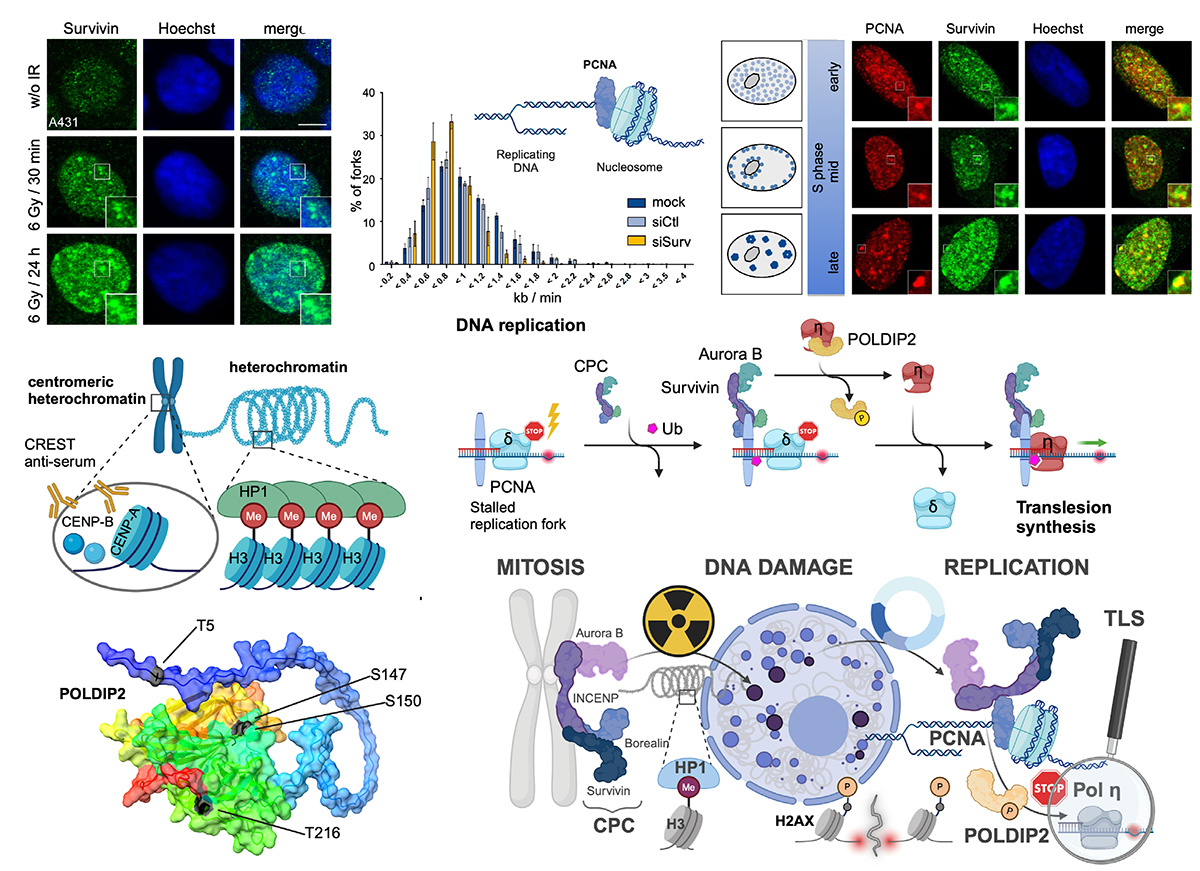
Survivin is known for its dual biological role in apoptosis inhibition and mitotic progression. In addition to its being part of the chromosomal passenger complex (CPC), recent findings suggest additional roles for Survivin in the DNA damage response, further contributing to therapy resistance. In this study, we investigated the role of Survivin and the CPC proteins in the cellular response to irradiation with a focus on DNA replication processes. As is known, ionizing radiation leads to an increased expression of Survivin and its accumulation in nuclear foci, which we now know to be specifically localized to centromeric heterochromatin. The depletion of Survivin and Aurora B increases the DNA damage marker γH2AX, indicative of an impaired repair capacity. The presence of Survivin and the CPC in nuclear foci that we already identified during the S phase co-localize with the proliferating cell nuclear antigen (PCNA), further implying a potential role during replication. Indeed, Survivin knockdown reduced replication fork speed as assessed via DNA fiber assays. Mechanistically, we identified a PIP-box motif in INCENP mediating the interaction with PCNA to assist in managing damage-induced replication stress. Survivin depletion forces cells to undergo unphysiological genome replication via mitotic DNA synthesis (MiDAS), resulting in chromosome breaks. Finally, we revealed that Aurora B kinase liberates Pol η by phosphorylating polymerase delta-interacting protein 2 (POLDIP2) to resume the replication of damaged sites via translesion synthesis. In this study, we assigned a direct function to the CPC in the transition from stalled replication forks to translesion synthesis, further emphasizing the ubiquitous overexpression of Survivin particularly in tumors. This study, for the first time, assigns a direct function to the chromosomal passenger complex, CPC, including Survivin, Aurora B kinase, Borealin, and INCENP, in the transition from stalled replication forks (involving PCNA binding) to translesion synthesis (liberating Pol η by phosphorylating POLDIP2), and thus in maintaining genomic integrity.
Tuning Nanobodies’ bioactivity against Survivin
Nanobodies are highly affine binders, often used to track disease-relevant proteins inside cells. However, they often fail to interfere with pathobiological functions, required for their clinical exploitation. Here, a nanobody targeting the disease-relevant apoptosis inhibitor and mitosis regulator Survivin (SuN) is utilized. Survivin’s multifaceted functions are regulated by an interplay of dynamic cellular localization, dimerization, and protein–protein interactions. However, as Survivin harbors no classical “druggable” binding pocket, one must aim at blocking extended protein surface areas. Comprehensive experimental evidence demonstrates that intracellular expression of SuN allows to track Survivin at low nanomolar concentrations but failed to inhibit its biological functions. Small angle X-ray scattering of the Survivin-SuN complex locates the proposed interaction interface between the C-terminus and the globular domain, as such not blocking any pivotal interaction. By clicking multiple SuN to ultrasmall (2 nm) gold nanoparticles (SuN-N), not only intracellular uptake is enabled, but additionally, Survivin crosslinking and interference with mitotic progression in living cells are also enabled. In sum, it is demonstrated that coupling of nanobodies to nanosized scaffolds can be universally applicable to improve their function and therapeutic applicability.
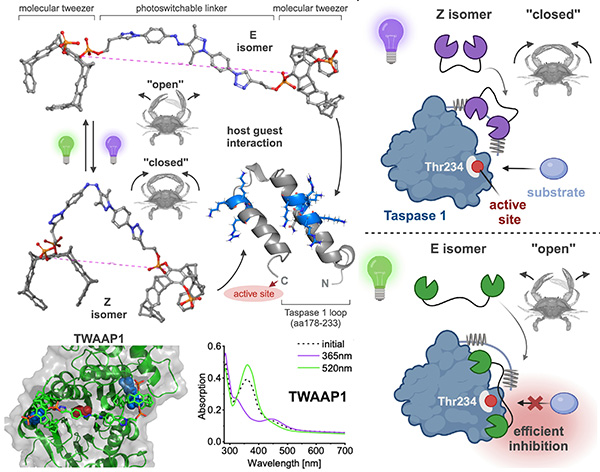
Photoresponsive molecular tweezers modulate Taspase 1 activity
Controlling protein activity with high precision in space and time remains a major challenge in chemical biology. In this study, we developed light-responsive molecular tweezers capable of dynamically modulating the activity of Taspase 1, a protease essential for embryogenesis and involved in tumor progression. These tweezers are designed to target lysine-rich regions within the flexible loop of Taspase 1, and incorporate photoswitchable arylazopyrazole (AAP) units that allow their binding geometry to be reversibly adjusted using visible light. The design relies on rigidly linking two molecular tweezers to the ends of the photoswitch, creating divalent ligands with precise and tunable spacing between the binding tips. Switching between the E and Z isomers alters the distance between the tweezers, enabling control over how the ligand engages the enzyme. Biochemical assays, surface plasmon resonance measurements, and molecular dynamics simulations demonstrated that these constructs bind Taspase 1 with high affinity and effectively inhibit its proteolytic activity. While both isomers could disrupt the interaction with the partner protein Importin α, the larger E-isomer tweezers exhibited particularly strong inhibition, likely due to their ability to bridge lysines flanking the active site. This photoswitchable tweezer system provides a powerful tool for precisely regulating protein function with light. By toggling the ligand’s configuration, researchers can selectively disrupt protein interactions, control enzymatic activity in real time, and study dynamic processes in complex biological environments. Beyond Taspase 1, this strategy offers a general approach for targeting other lysine-rich proteins, highlighting the potential of light-responsive supramolecular ligands for both fundamental research and the development of next-generation, precisely controllable therapeutics.
Here, too, one of our suggestions was accepted as the cover.
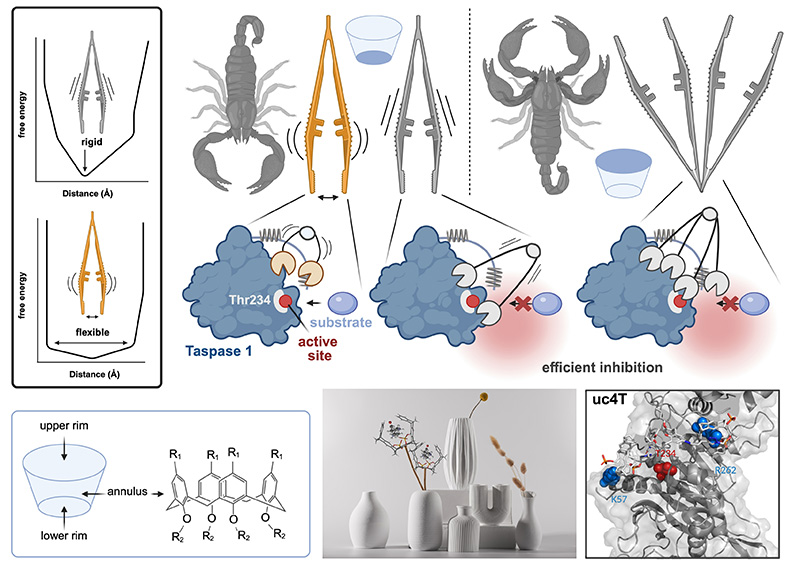
Highly Preorganized Calixarene Tweezers for Protein Recognition
Taspase 1 is a cancer-associated protease that plays a key role in cellular regulation and represents a challenging target for therapeutic intervention. In our study, we developed a series of specially designed calixarene-based molecular tweezers that are preorganized to interact precisely with Taspase 1. By carefully controlling the rigidity, valency, and arrangement of the binding units, these tweezers can selectively engage key residues within a flexible loop near the enzyme’s active site. We found that the most rigid and well-structured tweezers were the most effective at disrupting the interaction between Taspase 1 and its partner protein Importin α, thereby inhibiting the protease’s activity. Molecular simulations showed that these rigid constructs remain stably anchored at defined sites, in addition blocking access to the active site, whereas more flexible tweezers were less potent. Testing different modifications revealed that while polar groups improved solubility, increased multivalency and structural rigidity were critical for maximal inhibitory activity. Overall, our results demonstrate that highly preorganized calixarene tweezers act as potent allosteric inhibitors of Taspase 1. This study establishes a clear relationship between the tweezers’ structure and their functional performance, providing a blueprint for designing synthetic ligands that can selectively modulate proteolytic enzymes. Beyond Taspase 1, these findings highlight the potential of supramolecular scaffolds to target challenging protein surfaces and open new avenues for therapeutic and chemical biology applications.
Taspase1 facilitates Topoisomerase IIß-mediated DNA breaks driving estrogen-induced transcription
The human protease Taspase1 plays a pivotal role in developmental processes and cancerous diseases by processing critical regulators, such as the leukemia proto-oncoprotein MLL. Despite almost two decades of intense research, Taspase1's biology is, however, still poorly understood, and so far, its cellular function was not assigned to a superordinate biological pathway or a specific signaling cascade. Our data, gained by methods such as co-immunoprecipitation, LC- MS/MS and Topoisomerase II DNA cleavage assays, now functionally link Taspase1 and hormone- induced, Topoisomerase IIβ-mediated transient DNA double-strand breaks, leading to active transcription. The specific interaction with Topoisomerase IIα enhances the formation of DNA double-strand breaks that are a key prerequisite for stimulus-driven gene transcription. Moreover, Taspase1 alters the H3K4 epigenetic signature upon estrogen-stimulation by cleaving the chromatin-modifying enzyme MLL. As estrogen-driven transcription and MLL-derived epigenetic labelling are reduced upon Taspase1 siRNA-mediated knockdown, we finally characterize Taspase1 as a multi- functional co-activator of estrogen-stimulated transcription.
Funding
- Wilhelm Sander-Stiftung
"Dissecting Survivin (patho)biology by motional restriction of its flexible protein elements with dynamic supramolecular ligands", joint application with Prof. Michael Giese, Fakultät für Chemie (2025-2027) - José Carreras Stiftung
"PPROTAC-Nanobodies targeting Taspase 1 as precision therapeutics for the treatment of infant leukemias“ joint application with Dr. Mike Blüggel (2026-2027) - Brigitte und Dr. Konstanze Wegener-Stiftung
"Characterization and functionalization of Survivin-specific nanobodies for innovative cancer therapie" (2026-207) - Deutsche Forschungsgemeinschaft (DFG)
Joint Research
- DFG Research Training Group AMTEC-PRO
Advanced Methods and Technologies for Proton Therapy