Central carbohydrate metabolism (CCM) in hyperthermophilic Archaea “Focused Functional Genomics”

As an increasing amount of genome sequence information becomes available, one of the major challenges of the post-genome era is to elucidate the function of numerous hypothetical genes which were found in all studied genomes (20-40% of genes).
Genome circle Thermoproteus tenax.
Thermoproteus tenax became one of our model organisms, because of its ability for facultatively heterotrophic growth and its broad physiological versatility. The genome sequence of T. tenax has been deciphered in collaboration with R. Hensel, S. C. Schuster and H.-P. Klenk and the genome information was used for the reconstruction of the CCM pathways [17]. This project focused on the Emden-Meyerhof-Parnas (EMP) pathway, the Entner-Doudoroff (ED) pathway, citric acid cycle, glycogen and trehalose metabolism. We used this focused approach in order to integrate classical biochemical with new genomics-based methods and thus to fill the gap between sequence and function. This combination allowed us to i) identify missing links in the CCM, ii) elucidate functions for hypothetical proteins, and iii) to identify pathways that were supposed to be absent in hyperthermophiles and Archaea in general [17, 20]. Thus, we were able to demonstrate the presence of the semi-phosphorylative ED pathway in the hyperthermophile T. tenax and the thermoacidophile Sulfolobus solfataricus (collaboration with J. Van der Oost [17, 18, 20,25]) and the presence of the OtsA-OtsB pathway for trehalose synthesis [17] in Archaea.
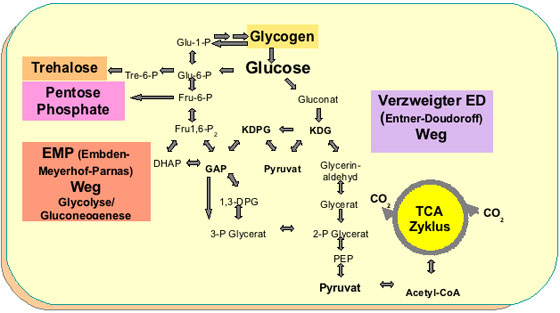
Central carbohydrate metabolism in Thermoproteus tenax.
