Success of Interdisciplinary Collaboration
Switching Off the Survival Protein for Cancer Cells
- 08.03.2021
It is called the "survival protein" because it plays a central role in the growth of cancer cells: survivin influences two important processes in the body's cells at the same time – cell death and cell division. Chemists and biologists at the UDE have now succeeded in developing a precise molecule that can bind the protein’s surface at a defined site and switch it off. "Nature Communications" covers the topic.*
Proteins control almost all vital processes in our cells. If they do not function correctly, if there is too much or too little of them, this can lead to the development of a variety of diseases including cancer. The associated proteins are therefore also important targets for drug discovery in biomedical research.
However, there are large number of proteins that simply do not offer suitable targets for a conventional active ingredient to dock onto. That's why scientists in UDE’s Collaborative Research Center 1093 are developing small, unusual molecules called supramolecular ligands that can precisely bind to their surface.
Molecules block pivotal site of the protein
Recently, the team of scientists led by Prof. Shirley Knauer, Prof. Elsa-Sanchez-Garcia and Prof. Thomas Schrader succeeded in targeting a critical interface that is important for the survival of cancer cells with such customized molecules. "The protein survivin is actually hardly found in healthy adult organisms," so Prof. Shirley Knauer. "In cancer cells, however, its production is ramped up." Using an artificial, tailored ligand, the scientists were able to cover the exact site of the survivin, which is responsible for its activation and transport out of the cell nucleus.
Each protein has a unique three-dimensional structure with a fissured surface that can form loops and niches. Sanchez-Garcia and her team performed computational analyses of the protein's surface and found out that the important interface is on an ordered but somewhat dynamic loop. Using this information and following further structural analyses, the chemists led by Schrader were able to design the ligand for this particularly difficult surface.
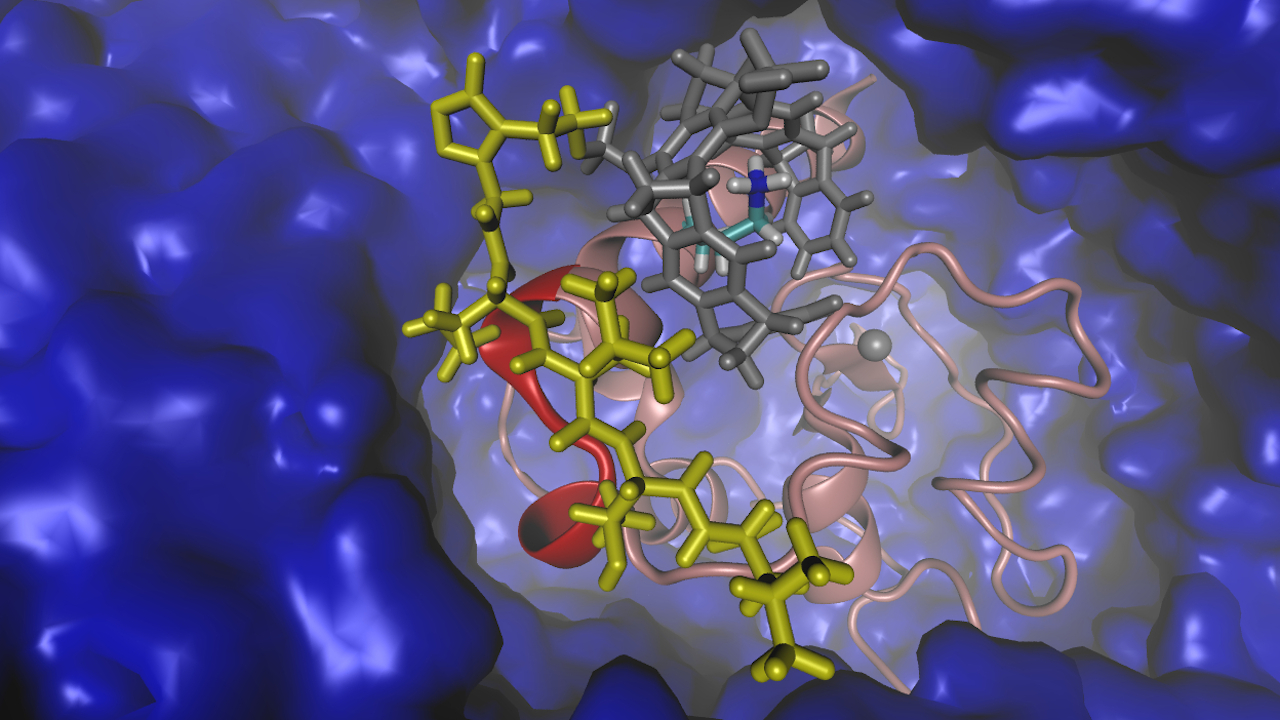
Image:
Graphical representation of the protein "Survivin" (red, pink) and the tailored ligand (gray, yellow) in water (blue).
* Publication: “Targeting a protein epitope: Specific inhibition of the Survivin-CRM1 interaction by peptide-modified molecular tweezers.”
Nature Communications 2021; https://doi.org/10.1038/s41467-021-21753-9
Further Information:
https://www.uni-due.de/crc1093/
Prof. Dr. Shirley Knauer, Molecular Biology II, +49 201/18 3-4987, shirley.knauer@uni-due.de
Editor: Dr. Lydia Didt, CRC 1093, +49 201/18 3-4351, lydia.didt@uni-due.de
